5JBN
 
 | |
3K86
 
 | |
5J1F
 
 | |
3K93
 
 | |
3JRR
 
 | | Crystal structure of the ligand binding suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 3 | | Authors: | Chan, J, Ishiyama, N, Ikura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 1.9 angstrom crystal structure of the suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor
To be Published
|
|
3JS5
 
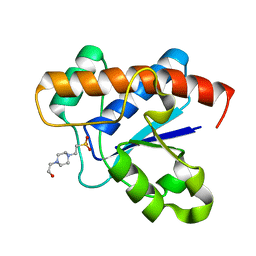 | |
3K9J
 
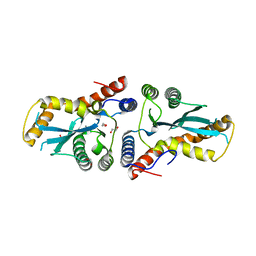 | | Transposase domain of Metnase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Histone-lysine N-methyltransferase SETMAR | | Authors: | Goodwin, K.D, He, H, Imasaki, T, Lee, S.-H, Georgiadis, M.M. | | Deposit date: | 2009-10-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of the human Hsmar1-derived transposase domain in the DNA repair enzyme Metnase.
Biochemistry, 49, 2010
|
|
3JSJ
 
 | |
5TO6
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Bandyopadhyay, A, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5JEA
 
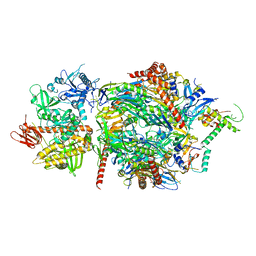 | | Structure of a cytoplasmic 11-subunit RNA exosome complex including Ski7, bound to RNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Exosome complex component CSL4, Exosome complex component MTR3, ... | | Authors: | Kowalinski, E, Ebert, J, Stegmann, E, Conti, E. | | Deposit date: | 2016-04-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex.
Mol.Cell, 63, 2016
|
|
3KA9
 
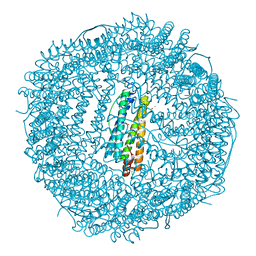 | | Frog M-ferritin, EEH mutant, with cobalt | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferritin, ... | | Authors: | Tosha, T, Ng, H.L, Theil, E, Alber, T, Bhattasali, O. | | Deposit date: | 2009-10-19 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Frog M-ferritin, EEH mutant, with cobalt
To be Published
|
|
3JV4
 
 | |
5J5J
 
 | | Crystal structure of a chimera of human Desmocollin-2 EC1 and human Desmoglein-2 EC2-EC5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2016-04-02 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis of adhesive binding by desmocollins and desmogleins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3K51
 
 | | Crystal Structure of DcR3-TL1A complex | | Descriptor: | Decoy receptor 3, Tumor necrosis factor ligand superfamily member 15, secreted form | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2009-10-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
6IMV
 
 | | The complex structure of endo-beta-1,2-glucanase from Talaromyces funiculosus with sophorose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanaka, N, Nakajima, M, Narukawa-Nara, M, Matsunaga, H, Kamisuki, S, Aramasa, H, Takahashi, Y, Sugimoto, N, Abe, K, Miyanaga, A, Yamashita, T, Sugawara, F, Kamakura, T, Komba, S, Nakai, H, Taguchi, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and structural analyses of a fungal endo-beta-1,2-glucanase reveal a new glycoside hydrolase family.
J.Biol.Chem., 294, 2019
|
|
2R0B
 
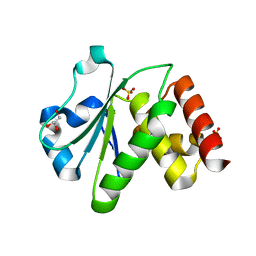 | | Crystal structure of human tyrosine phosphatase-like serine/threonine/tyrosine-interacting protein | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine/tyrosine-interacting protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-18 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
3J97
 
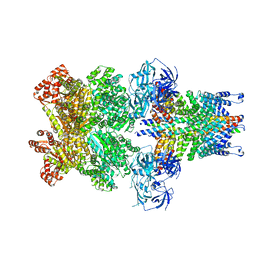 | | Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State II) | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Zhao, M, Wu, S, Cheng, Y, Brunger, A.T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanistic insights into the recycling machine of the SNARE complex.
Nature, 518, 2015
|
|
3K6T
 
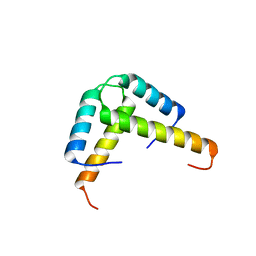 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans at 2.04 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-09 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
3K73
 
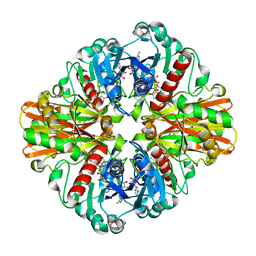 | | Crystal Structure of Phosphate bound Holo Glyceraldehyde-3-phosphate dehydrogenase 1 from MRSA252 at 2.5 Angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-10-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3JA1
 
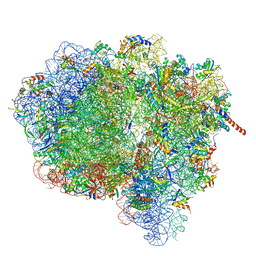 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
5THQ
 
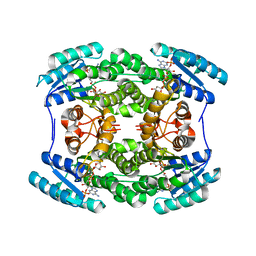 | |
6F25
 
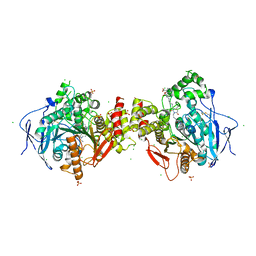 | | Crystal structure of human acetylcholinesterase in complex with C35. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dias, J, Nachon, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05199647 Å) | | Cite: | New evidence for dual binding site inhibitors of acetylcholinesterase as improved drugs for treatment of Alzheimer's disease.
Neuropharmacology, 155, 2019
|
|
5J91
 
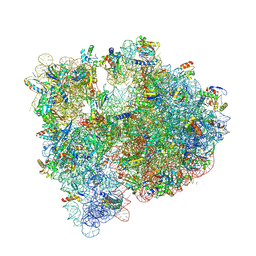 | | Structure of the Wild-type 70S E coli ribosome bound to Tigecycline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-07-06 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3JUE
 
 | | Crystal Structure of ArfGAP and ANK repeat domain of ACAP1 | | Descriptor: | ARFGAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | Authors: | Pang, X, Zhang, K, Ma, J, Zhou, Q, Sun, F. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
3JVT
 
 | | Calcium-bound Scallop Myosin Regulatory Domain (Lever Arm) with Reconstituted Complete Light Chains | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Himmel, D.M, Mui, S, O'Neall-Hennessey, E, Szent-Gyorgyi, A, Cohen, C. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The on-off switch in regulated myosins: different triggers but related mechanisms.
J.Mol.Biol., 394, 2009
|
|
