2JZF
 
 | | NMR Conformer closest to the mean coordinates of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
3EWP
 
 | | complex of substrate ADP-ribose with IBV Nsp3 ADRP domain | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
3EWO
 
 | | IBV Nsp3 ADRP domain | | Descriptor: | Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
2RNK
 
 | | NMR structure of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B.W, Wilson, I.A, Stevens, R.C, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
7WZO
 
 | |
3EWR
 
 | | complex of substrate ADP-ribose with HCoV-229E Nsp3 ADRP domain | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
3EWQ
 
 | | HCov-229E Nsp3 ADRP domain | | Descriptor: | Non-structural protein 3 | | Authors: | Xu, Y, Cong, L, Chen, C, Wei, L, Zhao, Q, Xu, X, Ma, Y, Bartlam, M, Rao, Z. | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two coronavirus ADP-ribose-1''-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain.
J.Virol., 83, 2009
|
|
7QG7
 
 | | SARS-CoV-2 macrodomain Nsp3b bound to the remdesivir nucleoside GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Wollenhaupt, J, Linhard, V, Sreeramulu, S, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
4OZA
 
 | | Backbone Modifications in the Protein GB1 Helix: beta-3-Ala24, beta-3-Lys28, beta-3-Gln32, beta-3-Asp36 | | Descriptor: | ISOPROPYL ALCOHOL, Streptococcal Protein GB1 Backbone Modified Variant: beta-3-Ala24, beta-3-Lys28, ... | | Authors: | Reinert, Z.E, Horne, W.S. | | Deposit date: | 2014-02-14 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Folding Thermodynamics of Protein-Like Oligomers with Heterogeneous Backbones.
Chem Sci, 5, 2014
|
|
6W02
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
8GQC
 
 | | Crystal structure of the SARS-unique domain (SUD) of SARS-CoV-2 (1.35 angstrom resolution) | | Descriptor: | Papain-like protease nsp3 | | Authors: | Qin, B, Li, Z, Aumonier, S, Wang, M, Cui, S. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of the SARS-unique domain of SARS-CoV-2 as an antiviral target.
Nat Commun, 14, 2023
|
|
8CMF
 
 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 nsp3 epitope (orf1ab)1350-1364 | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
8EUA
 
 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | Descriptor: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | Authors: | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | Deposit date: | 2022-10-18 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
5W6F
 
 | |
6MS9
 
 | | GDP-bound KRAS P34R mutant | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Bera, A.K, Westover, K.D. | | Deposit date: | 2018-10-16 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | GTP hydrolysis is modulated by Arg34 in the RASopathy-associated KRASP34R.
Birth Defects Res, 112, 2020
|
|
6O36
 
 | |
6O46
 
 | |
7XC3
 
 | |
7XC4
 
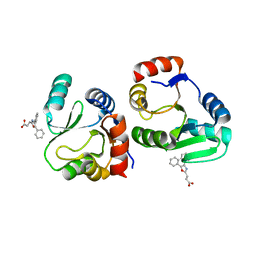 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain 3 (SARS-unique domain-M) in complex with Oxaprozin | | Descriptor: | 3-(4,5-diphenyl-1,3-oxazol-2-yl)propanoic acid, Papain-like protease nsp3 | | Authors: | Li, J, Liu, Y, Gao, J, Ruan, K. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Binding Sites of SARS-CoV-2 Macrodomain 3 Probed by Oxaprozin and Meclomen.
J.Med.Chem., 65, 2022
|
|
3T6G
 
 | |
3UJ4
 
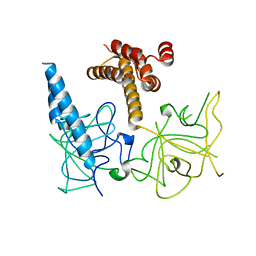 | | Crystal structure of the apo-inositol 1,4,5-trisphosphate receptor | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1, SULFATE ION | | Authors: | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
3UJ0
 
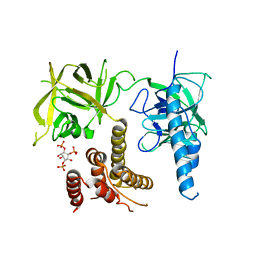 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor with ligand bound form. | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
3ETI
 
 | |
6MU1
 
 | | Structure of full-length IP3R1 channel bound with Adenophostin A | | Descriptor: | Adenophostin A, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Serysheva, I.I, Fan, G, Baker, M.R, Wang, Z, Seryshev, A, Ludtke, S.J, Baker, M.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM reveals ligand induced allostery underlying InsP3R channel gating.
Cell Res., 28, 2018
|
|
