7JXF
 
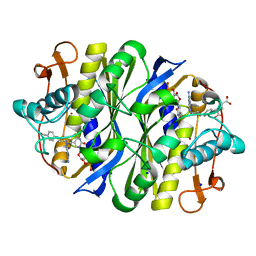 | | E. coli TSase complex with a bi-substrate reaction intermediate diastereomer analog | | Descriptor: | (2S)-2-({4-[({(6S)-2,4-diamino-5-[(1-{(2R,4S,5R)-4-hydroxy-5-[(phosphonooxy)methyl]tetrahydrofuran-2-yl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)methyl]-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl}methyl)amino]benzoyl}amino)pentanedioic acid (non-preferred name), 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE, N-[4-({[(6S)-2,4-diamino-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl]methyl}amino)benzene-1-carbonyl]-L-glutamic acid, ... | | Authors: | Finer-Moore, J, Kholodar, S.A, Stroud, R.M, Kohen, A, Moliner, V, Swiderek, K. | | Deposit date: | 2020-08-27 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caught in Action: X-ray Structure of Thymidylate Synthase with Noncovalent Intermediate Analog.
Biochemistry, 60, 2021
|
|
8TO8
 
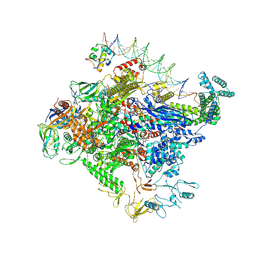 | | Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TOM
 
 | |
8TO1
 
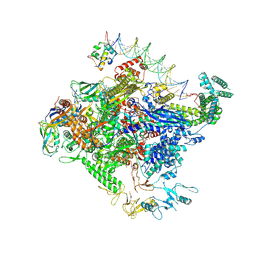 | | Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TOE
 
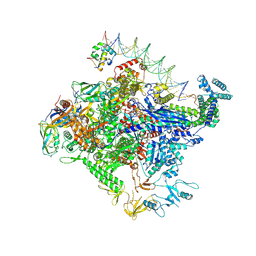 | |
8TO6
 
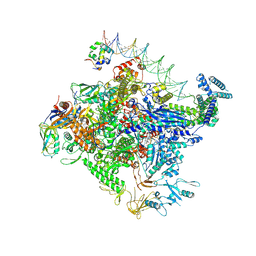 | | Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
6MR9
 
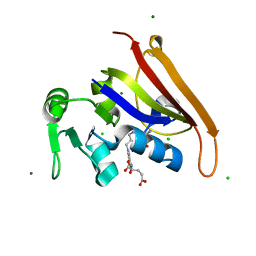 | | E. coli DHFR complex with a reaction intermediate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-10-12 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Time-resolved x-ray crystallography capture of a slow reaction tetrahydrofolate intermediate.
Struct Dyn., 6, 2019
|
|
5V84
 
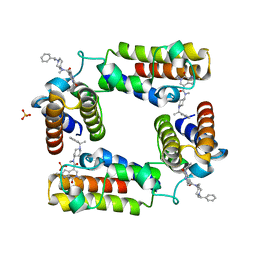 | | CECR2 in complex with Cpd6 (6-allyl-N,2-dimethyl-7-oxo-N-(1-(1-phenylethyl)piperidin-4-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4-carboxamide) | | Descriptor: | Cat eye syndrome critical region protein 2, N,2-dimethyl-7-oxo-N-{1-[(1S)-1-phenylethyl]piperidin-4-yl}-6-(prop-2-en-1-yl)-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-4- carboxamide, SULFATE ION | | Authors: | Murray, J.M, Kiefer, J.R, Jayaran, H, Bellon, S, Boy, F. | | Deposit date: | 2017-03-21 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | GNE-886: A Potent and Selective Inhibitor of the Cat Eye Syndrome Chromosome Region Candidate 2 Bromodomain (CECR2).
ACS Med Chem Lett, 8, 2017
|
|
7LS6
 
 | | Cryo-EM structure of Pre-15S proteasome core particle assembly intermediate purified from Pre3-1 proteasome mutant (G34D) | | Descriptor: | Proteasome assembly chaperone 2, Proteasome chaperone 1, Proteasome maturation factor UMP1, ... | | Authors: | Schnell, H.M, Walsh Jr, R.M, Rawson, S, Hanna, J.W. | | Deposit date: | 2021-02-17 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LSX
 
 | | Cryo-EM structure of 13S proteasome core particle assembly intermediate purified from Pre3-1 proteasome mutant (G34D) | | Descriptor: | Proteasome assembly chaperone 2, Proteasome chaperone 1, Proteasome maturation factor UMP1, ... | | Authors: | Schnell, H.M, Walsh Jr, R.M, Rawson, S, Hanna, J.W. | | Deposit date: | 2021-02-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7W5U
 
 | | Acetyl-CoA Carboxylase-AccB | | Descriptor: | Acetyl-CoA carboxylase complex, beta-chain, GLYCEROL, ... | | Authors: | Ali, I, Zheng, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of Acetyl-CoA carboxylase (AccB) from Streptomyces antibioticus and insights into the substrate-binding through in silico mutagenesis and biophysical investigations.
Comput Biol Med, 145, 2022
|
|
6RTM
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1-[(dimethylamino)methyl]-2-naphthol at 2 hour of soaking | | Descriptor: | 1-methylidenenaphthalen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Angelucci, F, Silvestri, I, Fata, F, Williams, D.L. | | Deposit date: | 2019-05-24 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ectopic suicide inhibition of thioredoxin glutathione reductase.
Free Radic Biol Med, 147, 2020
|
|
6RTJ
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1-[(dimethylamino)methyl]-2-naphthol at 1 hour of soaking | | Descriptor: | 1-[(dimethylamino)methyl]naphthalen-2-ol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Angelucci, F, Silvestri, I, Fata, F, Williams, D.L. | | Deposit date: | 2019-05-24 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ectopic suicide inhibition of thioredoxin glutathione reductase.
Free Radic Biol Med, 147, 2020
|
|
6RTO
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 1-[(dimethylamino)methyl]-2-naphthol at 4 hours of soaking | | Descriptor: | 1-methylidenenaphthalen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, TRIETHYLENE GLYCOL, ... | | Authors: | Angelucci, F, Silvestri, I, Fata, F, Williams, D.L. | | Deposit date: | 2019-05-24 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ectopic suicide inhibition of thioredoxin glutathione reductase.
Free Radic Biol Med, 147, 2020
|
|
5T4L
 
 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | (4R)-4-amino-6-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}hexanoic acid, Aspartate aminotransferase | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2LFL
 
 | | NMR solution structure of the intermediate IIIb of TdPI-short | | Descriptor: | Tryptase inhibitor | | Authors: | Bronsoms, S, Pantoja-Uceda, D, Gabrijelcic-Geiger, D, Sanglas, L, Aviles, F, Santoro, J, Sommerhoff, C, Arolas, J. | | Deposit date: | 2011-07-06 | | Release date: | 2011-11-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Oxidative folding and structural analyses of a kunitz-related inhibitor and its disulfide intermediates: functional implications.
J.Mol.Biol., 414, 2011
|
|
2LR3
 
 | |
7OQ6
 
 | | Crystal structure of cytochrome P450 Sas16 from Streptomyces asterosporus | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Zhang, L, Zhang, S, Bechthold, A, Einsle, O. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P450-mediated dehydrotyrosine formation during WS9326 biosynthesis proceeds via dehydrogenation of a specific acylated dipeptide substrate.
Acta Pharm Sin B, 13, 2023
|
|
1ETS
 
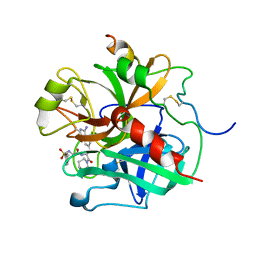 | |
4WHR
 
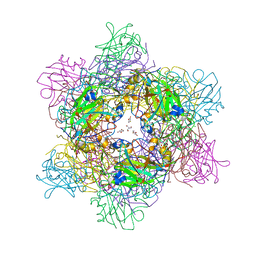 | | Anhydride reaction intermediate trapped in Protocatechuate 3,4-dioxygenase (pseudomonas putida) at pH 8.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-fluorobenzene-1,2-diol, 4-fluorooxepine-2,7-dione, ... | | Authors: | Knoot, C.J, Lipscomb, J.D. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of alkylperoxo and anhydride intermediates in an intradiol ring-cleaving dioxygenase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8GVA
 
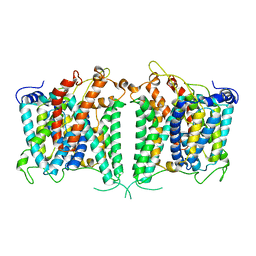 | | The intermediate structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
5VK2
 
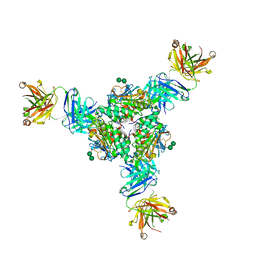 | | Structural basis for antibody-mediated neutralization of Lassa virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hastie, K.M, Zandonatti, M.A, Kleinfelter, L.M, Rowland, M.L, Rowland, M.M, Chandra, K, Branco, L.M, Robinson, J.E, Garry, R.F, Saphire, E.O. | | Deposit date: | 2017-04-20 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis for antibody-mediated neutralization of Lassa virus.
Science, 356, 2017
|
|
2YKA
 
 | | RRM domain of mRNA export adaptor REF2-I bound to HVS ORF57 peptide | | Descriptor: | 52 KDA IMMEDIATE-EARLY PHOSPHOPROTEIN, RNA AND EXPORT FACTOR-BINDING PROTEIN 2 | | Authors: | Tunnicliffe, R.B, Hautbergue, G.M, Kalra, P, Wilson, S.A, Golovanov, A.P. | | Deposit date: | 2011-05-26 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri Orf57 to the Mammalian Export Adaptor Alyref.
Plos Pathog., 10, 2014
|
|
5WT4
 
 | | L-Cysteine-PLP intermediate of NifS from Helicobacter pylori | | Descriptor: | Cysteine desulfurase IscS, ISOPROPYL ALCOHOL, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Fujishiro, T, Nakamura, R, Takahashi, T. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 2019
|
|
1BVV
 
 | |
