9DCH
 
 | | Single-stranded RNA-mediated PRC2 dimer | | Descriptor: | Isoform 2 of Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, Polycomb protein SUZ12, ... | | Authors: | Song, J.S, Kasinath, V.K. | | Deposit date: | 2024-08-26 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Diverse RNA Structures Induce PRC2 Dimerization and Inhibit Histone Methyltransferase Activity.
Biorxiv, 2024
|
|
9C8U
 
 | | Human PRC2 - RvLEAM (short) (1:6 molar ratio), cross-linked 10 min | | Descriptor: | Isoform 2 of Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, Polycomb protein SUZ12, ... | | Authors: | Abe, K.M, Li, G, He, Q, Grant, T, Lim, C. | | Deposit date: | 2024-06-13 | | Release date: | 2024-09-04 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Small LEA proteins mitigate air-water interface damage to fragile cryo-EM samples during plunge freezing.
Nat Commun, 15, 2024
|
|
1AP0
 
 | | STRUCTURE OF THE CHROMATIN BINDING (CHROMO) DOMAIN FROM MOUSE MODIFIER PROTEIN 1, NMR, 26 STRUCTURES | | Descriptor: | MODIFIER PROTEIN 1 | | Authors: | Ball, L.J, Murzina, N.V, Broadhurst, R.W, Raine, A.R.C, Archer, S.J, Stott, F.J, Murzin, A.G, Singh, P.B, Domaille, P.J, Laue, E.D. | | Deposit date: | 1997-07-22 | | Release date: | 1998-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the chromatin binding (chromo) domain from mouse modifier protein 1.
EMBO J., 16, 1997
|
|
3QO2
 
 | | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9 | | Descriptor: | 1,2-ETHANEDIOL, Histone H3 peptide, M-phase phosphoprotein 8 | | Authors: | Chang, Y, Horton, J.R, Bedford, M.T, Zhang, X, Cheng, X. | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding.
J.Mol.Biol., 408, 2011
|
|
7VYW
 
 | |
7VZ2
 
 | | Crystal structure of chromodomain of Arabidopsis LHP1 | | Descriptor: | Chromo domain-containing protein LHP1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Min, J. | | Deposit date: | 2021-11-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the recognition of methylated histone H3 by the Arabidopsis LHP1 chromodomain.
J.Biol.Chem., 298, 2022
|
|
8Z7D
 
 | | Structure of G9a in complex with compound 9a | | Descriptor: | 3,6,6-trimethyl-4-oxidanylidene-~{N}-[(2~{S})-1-oxidanylidene-1-[(phenylmethyl)amino]hexan-2-yl]-5,7-dihydro-1~{H}-indole-2-carboxamide, Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-04-20 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structure-based development of novel substrate-type G9a inhibitors as epigenetic modulators for sickle cell disease treatment.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
8Z7E
 
 | | Structure of G9a in complex with compound 9b | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ZINC ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-04-20 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Structure-based development of novel substrate-type G9a inhibitors as epigenetic modulators for sickle cell disease treatment.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
8Z7C
 
 | | Structure of G9a in complex with compound 7i | | Descriptor: | 3,6,6-trimethyl-~{N}-[(2~{S})-1-[[4-(1-methylpiperidin-4-yl)oxyphenyl]amino]-1-oxidanylidene-hexan-2-yl]-4-oxidanylidene-5,7-dihydro-1~{H}-indole-2-carboxamide, Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-04-20 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-based development of novel substrate-type G9a inhibitors as epigenetic modulators for sickle cell disease treatment.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
8EQV
 
 | | Cryo-EM structure of PRC2 in complex with the long isoform of AEBP2 | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | Authors: | Boudes, M, Zhang, Q, Flanigan, S.F, Davidovich, C. | | Deposit date: | 2022-10-09 | | Release date: | 2024-07-31 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | To be updated
To Be Published
|
|
8F8Y
 
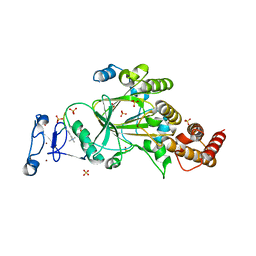 | | PHF2 (PHD+JMJ) in Complex with VRK1 N-Terminal Peptide | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase PHF2, SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2022-11-22 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A complete methyl-lysine binding aromatic cage constructed by two domains of PHF2.
J.Biol.Chem., 299, 2022
|
|
8F8Z
 
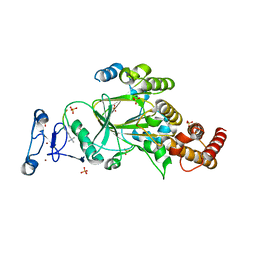 | |
7KSO
 
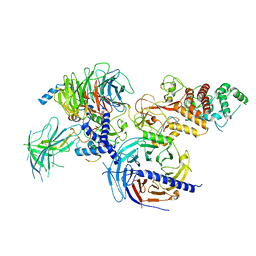 | | Cryo-EM structure of PRC2:EZH1-AEBP2-JARID2 | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
7KSR
 
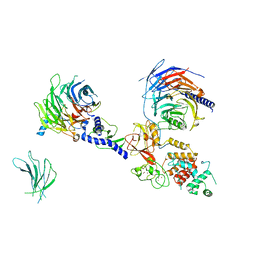 | | PRC2:EZH1_A from a dimeric PRC2 bound to a nucleosome | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
7KTP
 
 | | PRC2:EZH1_B from a dimeric PRC2 bound to a nucleosome | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
4IUV
 
 | |
4IUU
 
 | |
6D08
 
 | | Crystal structure of an engineered bump-hole complex of mutant human chromobox homolog 1 (CBX1) with H3K9bn peptide | | Descriptor: | Chromobox protein homolog 1, GLYCEROL, Histone H3.1, ... | | Authors: | Arora, S, Horne, W.S, Islam, K. | | Deposit date: | 2018-04-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering Methyllysine Writers and Readers for Allele-Specific Regulation of Protein-Protein Interactions.
J.Am.Chem.Soc., 141, 2019
|
|
8UXQ
 
 | |
6D07
 
 | |
8VML
 
 | | PRC2_AJ1-450 bound to H3K4me3 | | Descriptor: | AEPB2, EED, EZH2, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VNZ
 
 | |
8VMI
 
 | | PRC2_AJ119-450 bound to H3K4me3 | | Descriptor: | EZH2, Histone H3.1, Histone H3.1t, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6C23
 
 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-05 | | Release date: | 2018-01-24 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
6C24
 
 | | Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, JARID2-substrate, ... | | Authors: | Kasinath, V, Faini, M, Poepsel, S, Reif, D, Feng, A, Stjepanovic, G, Aebersold, R, Nogales, E. | | Deposit date: | 2018-01-06 | | Release date: | 2018-01-24 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Science, 359, 2018
|
|
