3L95
 
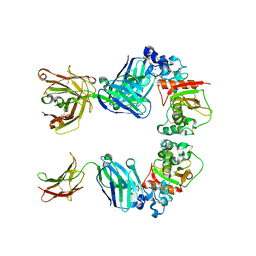 | |
3PSC
 
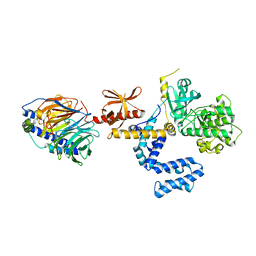 | | Bovine GRK2 in complex with Gbetagamma subunits | | Descriptor: | Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Thal, D.M, Tesmer, J.J. | | Deposit date: | 2010-12-01 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Molecular Mechanism of Selectivity among G Protein-Coupled Receptor Kinase 2 Inhibitors.
Mol.Pharmacol., 80, 2011
|
|
4BUJ
 
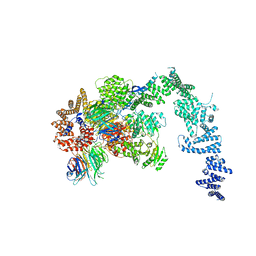 | | Crystal structure of the S. cerevisiae Ski2-3-8 complex | | Descriptor: | ANTIVIRAL HELICASE SKI2, ANTIVIRAL PROTEIN SKI8, SULFATE ION, ... | | Authors: | Halbach, F, Reichelt, P, Rode, M, Conti, E. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The Yeast Ski Complex: Crystal Structure and RNA Channeling to the Exosome Complex.
Cell(Cambridge,Mass.), 154, 2013
|
|
4BZJ
 
 | | The structure of the COPII coat assembled on membranes | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
4Z28
 
 | | Crystal structure of short hoefavidin biotin complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Avidin family, BIOTIN | | Authors: | Livnah, O, Avraham, O. | | Deposit date: | 2015-03-29 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Hoefavidin: A dimeric bacterial avidin with a C-terminal binding tail.
J.Struct.Biol., 191, 2015
|
|
4Z6J
 
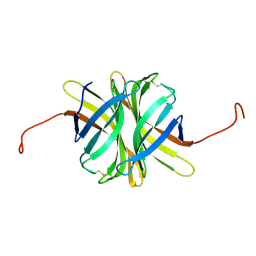 | |
6S13
 
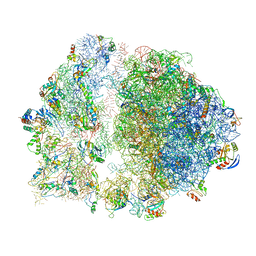 | | Erythromycin Resistant Staphylococcus aureus 70S ribosome (delta R88 A89 uL22). | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Halfon, Y, Matozv, D, Eyal, Z, Bashan, A, Zimmerman, E, Kjeldgaard, J, Ingmer, H, Yonath, A. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant.
Sci Rep, 9, 2019
|
|
4GGD
 
 | |
6RLQ
 
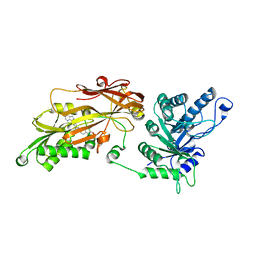 | | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJ45031882 | | Descriptor: | (1~{S},2~{R},3~{S},5~{R})-3-[2-(2-azanyl-3-bromanyl-quinolin-7-yl)ethyl]-5-(4-azanylpyrrolo[2,3-d]pyrimidin-7-yl)cyclop entane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Brown, D.G, Robinson, C.M, Pande, V. | | Deposit date: | 2019-05-02 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJ45031882
To Be Published
|
|
6RM3
 
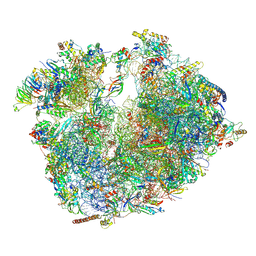 | | Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | Authors: | Barandun, J, Hunziker, M, Vossbrinck, C.R, Klinge, S. | | Deposit date: | 2019-05-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome.
Nat Microbiol, 4, 2019
|
|
4CY1
 
 | | Crystal structure of the KANSL1-WDR5 complex. | | Descriptor: | GLYCEROL, KAT8 REGULATORY NSL COMPLEX SUBUNIT 1, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
4CGB
 
 | | Crystal structure of the trimerization domain of EML2 | | Descriptor: | ECHINODERM MICROTUBULE-ASSOCIATED PROTEIN-LIKE 2, GLYCEROL, POTASSIUM ION | | Authors: | Richards, M.W, Bayliss, R. | | Deposit date: | 2013-11-21 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Microtubule Association of Eml Proteins and the Eml4-Alk Variant 3 Oncoprotein Require an N-Terminal Trimerization Domain.
Biochem.J., 467, 2015
|
|
6S1L
 
 | |
4GM8
 
 | | Crystal structure of human WD repeat domain 5 with compound MM-102 | | Descriptor: | MM-102, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction.
J.Am.Chem.Soc., 135, 2013
|
|
6RXY
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state a | | Descriptor: | 35S rRNA, 40S ribosomal protein S13-like protein, 40S ribosomal protein S14-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXT
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state A | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6S1R
 
 | |
4CY2
 
 | | Crystal structure of the KANSL1-WDR5-KANSL2 complex. | | Descriptor: | KAT8 REGULATORY NSL COMPLEX SUBUNIT 1, KAT8 REGULATORY NSL COMPLEX SUBUNIT 2, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
3HAZ
 
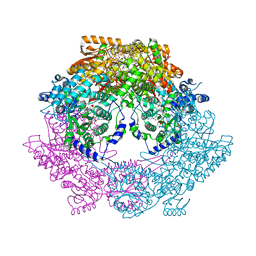 | | Crystal structure of bifunctional proline utilization A (PutA) protein | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2009-05-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4CY3
 
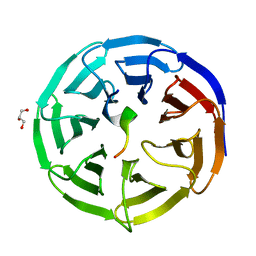 | | Crystal structure of the NSL1-WDS complex. | | Descriptor: | CG4699, ISOFORM D, GLYCEROL, ... | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
4GQB
 
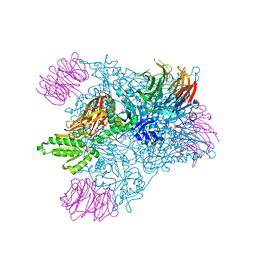 | | Crystal Structure of the human PRMT5:MEP50 Complex | | Descriptor: | (2S,5S,6E)-2,5-diamino-6-[(3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxydihydrofuran-2(3H)-ylidene]hexanoic acid, Histone H4 peptide, Methylosome protein 50, ... | | Authors: | Antonysamy, S, Bonday, Z, Campbell, R, Doyle, B, Druzina, Z, Gheyi, T, Han, B, Jungheim, L.N, Qian, Y, Rauch, C, Russell, M, Sauder, J.M, Wasserman, S.R, Weichert, K, Willard, F.S, Zhang, A, Emtage, S. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the human PRMT5:MEP50 complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8P16
 
 | | E167K RF2 on E. coli 70S release complex with UGG (Structure I) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Pundir, S, Larsson, D.S.D, Selmer, M, Sanyal, S. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | The compensatory mechanism of a naturally evolved
E167K RF2 counteracting the loss of
RF1 in bacteria
To Be Published
|
|
4CY5
 
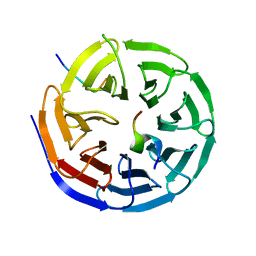 | | Crystal structure of the NSL1-WDS-NSL2 complex. | | Descriptor: | CG4699, ISOFORM D, DIM GAMMA-TUBULIN 1, ... | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
6S0X
 
 | | Erythromycin Resistant Staphylococcus aureus 70S ribosome (delta R88 A89 uL22) in complex with erythromycin. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Halfon, Y, Matozv, D, Eyal, Z, Bashan, A, Zimmerman, E, Kjeldgaard, J, Ingmer, H, Yonath, A. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (2.425 Å) | | Cite: | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant.
Sci Rep, 9, 2019
|
|
6RXV
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B2 | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
