4Z41
 
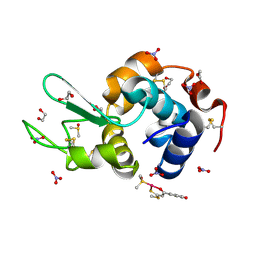 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Compound with a S,O Bidentate Ligand (9a=Chloro-(1-(3'-hydroxy)-3-(methylthio)-3-thioxo-prop-1-en-1-olate-O,S)-(dimethylsulfoxide-S)-platinum(II)) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A. | | Deposit date: | 2015-04-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
6ER4
 
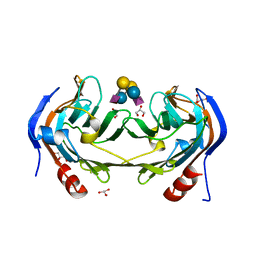 | | Ruminococcus gnavus IT-sialidase CBM40 bound to alpha2,6 sialyllactose | | Descriptor: | BNR/Asp-box repeat protein, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2017-10-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus.
Nat Commun, 8, 2017
|
|
6ELO
 
 | |
8PBB
 
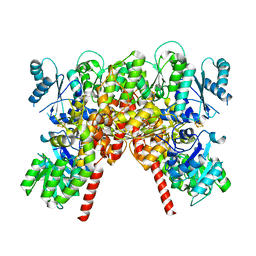 | | CHAPSO treated partial catalytic component (comprising only AnfD & AnfK, lacking AnfG and FeFeco) of iron nitrogenase from Rhodobacter capsulatus | | Descriptor: | FE(8)-S(7) CLUSTER, Nitrogenase iron-iron protein, beta subunit, ... | | Authors: | Schmidt, F.V, Schulz, L, Zarzycki, J, Prinz, S, Erb, T.J, Rebelein, J.G. | | Deposit date: | 2023-06-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural insights into the iron nitrogenase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3H1G
 
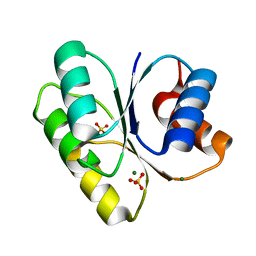 | |
8P52
 
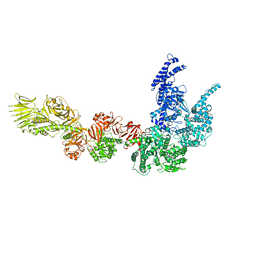 | |
6E6A
 
 | | Triclinic crystal form of IncA G144A point mutant | | Descriptor: | Inclusion membrane protein A, SODIUM ION | | Authors: | Cingolani, G, Paumet, F. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the homotypic fusion of chlamydial inclusions by the SNARE-like protein IncA.
Nat Commun, 10, 2019
|
|
8P51
 
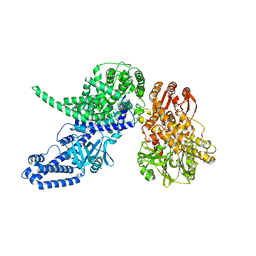 | |
3GLZ
 
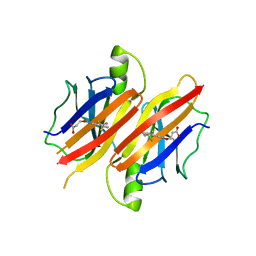 | | Human Transthyretin (TTR) complexed with(E)-3-(2-(trifluoromethyl)benzylideneaminooxy)propanoic acid (inhibitor 11) | | Descriptor: | 3-[({(1E)-[2-(trifluoromethyl)phenyl]methylidene}amino)oxy]propanoic acid, Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Orlandini, E, Sacchettini, J.C. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis
Plos One, 4, 2009
|
|
6YE9
 
 | | Small-molecule inhibitor of 14-3-3 protein-protein interactions | | Descriptor: | 14-3-3 protein sigma, MAGNESIUM ION, [2-[2-oxidanylidene-2-[(phenylmethyl)amino]ethoxy]phenyl]phosphonic acid | | Authors: | Ottmann, C, Visser, E.J. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based conversion of a promiscuous inhibitor to a selective stabilizer of protein-protein interactions
To Be Published
|
|
8P50
 
 | | Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin with the C-terminal deletion in complex with Arf3 | | Descriptor: | ADP-ribosylation factor 3, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Belyy, A, Heilen, P, Hofnagel, O, Raunser, S. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structure and activation mechanism of the Makes caterpillars floppy 1 toxin.
Nat Commun, 14, 2023
|
|
6ELN
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 4-[4-(4-methoxyphenyl)-5-methyl-1H-pyrazol-3-yl]benzene-1,3-diol, Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-09-29 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
8ZLG
 
 | |
8ZMC
 
 | | Dengue 3 NS5 methyltransferase bound to S-Adenosyl-L-homocysteine and Herbacetin | | Descriptor: | Dengue 3 NS5 methyltransferase, GLYCEROL, Herbacetin, ... | | Authors: | Bhutkar, M, Verma, S, Tomar, S, Kumar, P. | | Deposit date: | 2024-05-23 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dengue 3 NS5 methyltransferase bound to S-Adenosyl-L-homocysteine and Herbacetin
To Be Published
|
|
6YRI
 
 | |
4Z0B
 
 | | Crystal Structure of the Fab Fragment of Anti-ofloxacin Antibody and Exploration Its Receptor Binding Site | | Descriptor: | PHOSPHATE ION, antibody heavy chain, antibody light chain | | Authors: | He, K, Du, X, Sheng, W, Zhou, X, Wang, J, Wang, S. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Fab Fragment of an Anti-ofloxacin Antibody and Exploration of Its Specific Binding.
J.Agric.Food Chem., 64, 2016
|
|
8PJN
 
 | | Catalytic module of human CTLH E3 ligase bound to multiphosphorylated UBE2H~ubiquitin | | Descriptor: | E3 ubiquitin-protein transferase MAEA, E3 ubiquitin-protein transferase RMND5A, Ubiquitin, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | Deposit date: | 2023-06-23 | | Release date: | 2024-01-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
6ER2
 
 | |
8PMQ
 
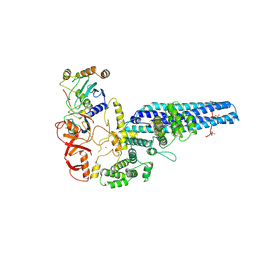 | | Catalytic module of yeast GID E3 ligase bound to multiphosphorylated Ubc8~ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, Ubiquitin, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
6E6C
 
 | | HRAS G13D bound to GppNHp (H13GNP) | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
6E6H
 
 | | NRAS G13D bound to GppNHp (N13GNP) | | Descriptor: | GTPase NRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
8PD0
 
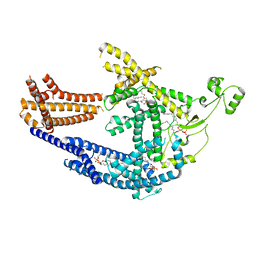 | | cryo-EM structure of Doa10 in MSP1E3D1 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | Authors: | Botsch, J.J, Braeuning, B, Schulman, B.A. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
4Z3M
 
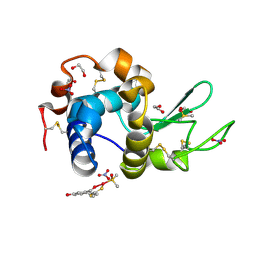 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Complex with S,O Bidentate Ligands (9b) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A. | | Deposit date: | 2015-03-31 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
8PDA
 
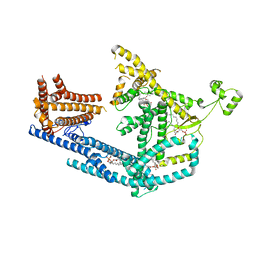 | | cryo-EM structure of Doa10 with RING domain in MSP1E3D1 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | Authors: | Botsch, J.J, Braeuning, B, Schulman, B.A. | | Deposit date: | 2023-06-12 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
6ELK
 
 | | C.elegans MnSOD-3 mutant - Q142H | | Descriptor: | GLYCEROL, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hunter, G.J, Trinh, C.H, Hunter, T. | | Deposit date: | 2017-09-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Single Mutation is Sufficient to Modify the Metal Selectivity and Specificity of a Eukaryotic Manganese Superoxide Dismutase to Encompass Iron.
Chemistry, 24, 2018
|
|
