6JM9
 
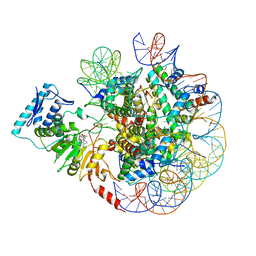 | | cryo-EM structure of DOT1L bound to unmodified nucleosome | | Descriptor: | DNA strand I, DNA strand J, Histone H2A, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
8VLR
 
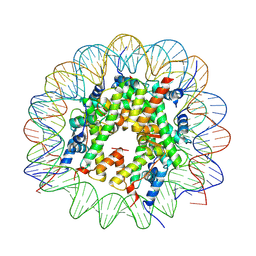 | | Cryo-EM structure of native H2AK119bu nucleosome at 2.6 | | Descriptor: | DNA (136-MER), Histone H2A type 1-B/E, Histone H2B type 1-A, ... | | Authors: | Wang, Y, Zhang, K. | | Deposit date: | 2024-01-12 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Role of histone variants H2BC1 and H2AZ.2 in H2AK119ub nucleosome organization and Polycomb gene silencing
To Be Published
|
|
6JMA
 
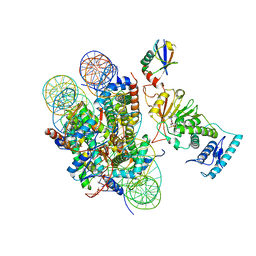 | | cryo-EM structure of DOT1L bound to H2B ubiquitinated nucleosome | | Descriptor: | DNA I&J, Histone H2A, Histone H2B 1.1, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
8WG5
 
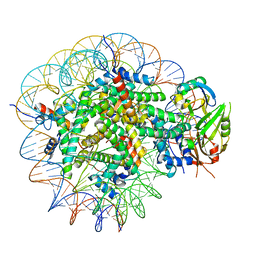 | | Cryo-EM structure of USP16 bound to H2AK119Ub nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, He, Z.Z, Deng, Z.H, Liu, L. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural and mechanistic basis for nucleosomal H2AK119 deubiquitination by single-subunit deubiquitinase USP16.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6K0C
 
 | | Crystal structure of ceNAP1-H2A.Z-H2B complex | | Descriptor: | Histone H2B 2,Histone H2A.V, Nucleosome Assembly Protein | | Authors: | Liu, Y.R. | | Deposit date: | 2019-05-05 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Crystal structure of xlH2A-H2B
Structure, 2019
|
|
8WHA
 
 | | Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH5
 
 | | Structure of DDM1-nucleosome complex in the apo state | | Descriptor: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHB
 
 | | Structure of nucleosome core particle of Arabidopsis thaliana | | Descriptor: | DNA (antisense strand), DNA (sense strand), Histone H2A.6, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-23 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH8
 
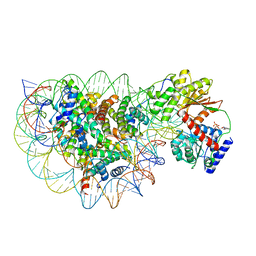 | | Structure of DDM1-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, DNA (antisense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
6HKT
 
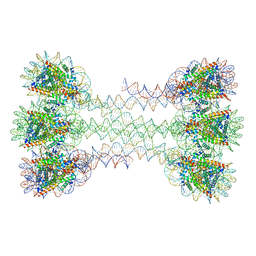 | | Structure of an H1-bound 6-nucleosome array | | Descriptor: | DNA (1122-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Garcia-Saez, I, Dimitrov, S, Petosa, C. | | Deposit date: | 2018-09-08 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (9.7 Å) | | Cite: | Structure of an H1-Bound 6-Nucleosome Array Reveals an Untwisted Two-Start Chromatin Fiber Conformation.
Mol. Cell, 72, 2018
|
|
8W9F
 
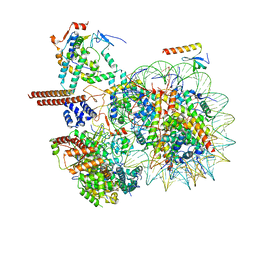 | |
6K03
 
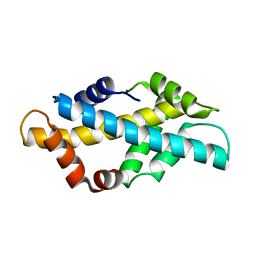 | | Crystal structure of ceH2A-H2B | | Descriptor: | Histone H2B 1,Histone H2A | | Authors: | Liu, Y.R. | | Deposit date: | 2019-05-05 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.857 Å) | | Cite: | Crystal structure of xlH2A-H2B
Structure, 2019
|
|
6K00
 
 | | Crystal structure A of ceNAP1-H2A-H2B complex | | Descriptor: | Histone H2B 1,Histone H2A, Nucleosome Assembly Protein | | Authors: | Liu, Y.R. | | Deposit date: | 2019-05-05 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure of xlH2A-H2B
Structure, 2019
|
|
6JXD
 
 | |
6I84
 
 | | Structure of transcribing RNA polymerase II-nucleosome complex | | Descriptor: | DNA (158-MER), DNA (170-MER), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Farnung, L, Vos, S.M, Cramer, P. | | Deposit date: | 2018-11-19 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of transcribing RNA polymerase II-nucleosome complex.
Nat Commun, 9, 2018
|
|
8VX6
 
 | | Human OGG1 bound at the nucleosomal DNA entry site | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | You, Q, Li, H. | | Deposit date: | 2024-02-03 | | Release date: | 2024-10-09 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human 8-oxoguanine glycosylase OGG1 binds nucleosome at the dsDNA ends and the super-helical locations.
Commun Biol, 7, 2024
|
|
8VX5
 
 | | Nucleosome core particle containing an 8-oxoG damage site | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | You, Q, Li, H. | | Deposit date: | 2024-02-03 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Human 8-oxoguanine glycosylase OGG1 binds nucleosome at the dsDNA ends and the super-helical locations.
Commun Biol, 7, 2024
|
|
8VWT
 
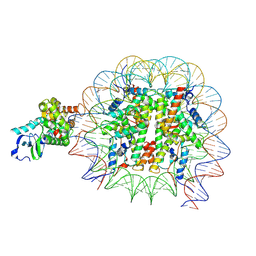 | | OGG1 bound to a nucleosome containing 8oxoG at SHL-6 (composite map) | | Descriptor: | 601 I strand (non-damaged strand), 601 J strand (damaged strand), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8VWU
 
 | | Nucleosome containing 8oxoG at SHL4 | | Descriptor: | 601 I strand (damaged strand), 601 J strand (non-damaged strand), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
6INQ
 
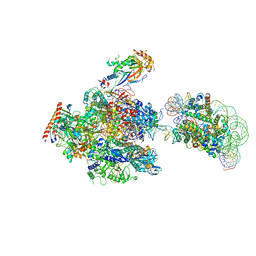 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA (+1 position) | | Descriptor: | DNA (198-MER), DNA (31-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-10-26 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6IQ4
 
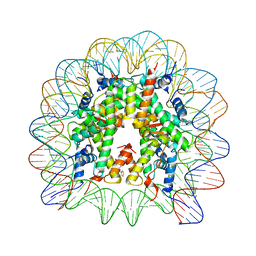 | | Nucleosome core particle cross-linked with a hetero-binuclear molecule possessing RAPTA and gold(I) 4-(diphenylphosphino)benzoic acid groups. | | Descriptor: | 4-diphenylphosphanylbenzoic acid, DNA (145-MER), GOLD ION, ... | | Authors: | DeFalco, L, Batchelor, L.K, Adhireksan, Z, Dyson, P.J, Davey, C.A. | | Deposit date: | 2018-11-06 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crosslinking Allosteric Sites on the Nucleosome.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8VWV
 
 | | OGG1 bound to a nucleosome containing 8oxoG at SHL4 (composite map) | | Descriptor: | 601 I strand (damaged strand), 601 J strand (non-damaged), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8VWS
 
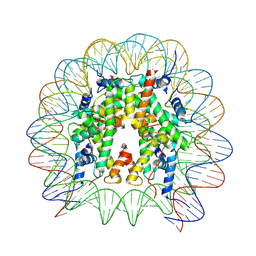 | | Nucleosome containing 8oxoG at SHL-6 | | Descriptor: | 601 I strand (non-damaged strand), 601 J strand (damaged strand), Histone H2A type 1, ... | | Authors: | Weaver, T.M, Ling, J.A, Freudenthal, B.D. | | Deposit date: | 2024-02-02 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Contributing factors to the oxidation-induced mutational landscape in human cells.
Nat Commun, 15, 2024
|
|
8VMJ
 
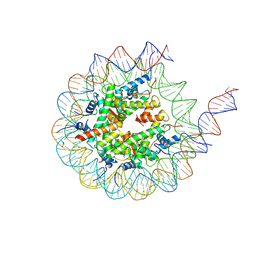 | | H3K4me3 nucleosome bound to PRC2_AJ119-450 | | Descriptor: | DNA (157-MER), Histone H2A, Histone H2B, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VOB
 
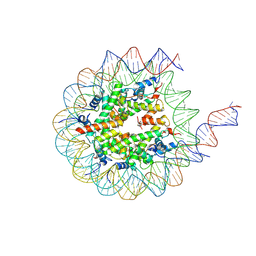 | | H3K36me3-modified nucleosome bound to PRC2_AJ1-450 | | Descriptor: | DNA (157-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-14 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
