1Q3H
 
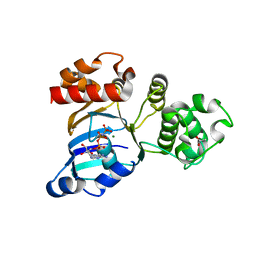 | | mouse CFTR NBD1 with AMP.PNP | | Descriptor: | ACETIC ACID, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, ... | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
2OQI
 
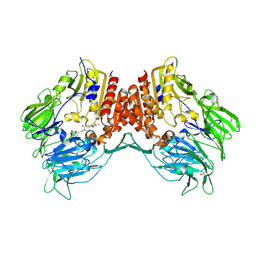 | | Human Dipeptidyl Peptidase IV (DPP4) with Piperidinone-constrained phenethylamine | | Descriptor: | (4R,5R)-5-AMINO-1-[2-(1,3-BENZODIOXOL-5-YL)ETHYL]-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-2-ONE, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) | | Authors: | Pei, Z, Li, X, von Geldern, T.W, Longenecker, K.L, Pireh, D, Stewart, K.D, Backes, B.J, Lai, C, Lubben, T.H, Ballaron, S.J, Beno, D.W, Kempf-Grote, A.J, Sham, H.L, Trevillyan, J.M. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of Piperidinone- and Piperidine-Constrained Phenethylamines as Novel, Potent, and Selective Dipeptidyl Peptidase IV Inhibitors.
J.Med.Chem., 50, 2007
|
|
4ITP
 
 | | Structure of human carbonic anhydrase II bound to a benzene sulfonamide | | Descriptor: | 2-phenyl-N-(4-sulfamoylbenzyl)acetamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Biswas, S, McKenna, R. | | Deposit date: | 2013-01-18 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Structural study of the location of the phenyl tail of benzene sulfonamides and the effect on human carbonic anhydrase inhibition.
Bioorg.Med.Chem., 21, 2013
|
|
2OVX
 
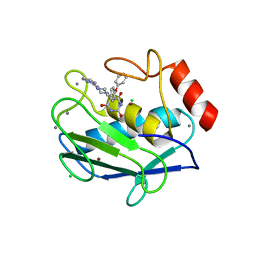 | | MMP-9 active site mutant with barbiturate inhibitor | | Descriptor: | 5-(4-PHENOXYPHENYL)-5-(4-PYRIMIDIN-2-YLPIPERAZIN-1-YL)PYRIMIDINE-2,4,6(2H,3H)-TRIONE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tochowicz, A, Bode, W, Maskos, K, Goettig, P. | | Deposit date: | 2007-02-15 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-chain to Selectivity.
J.Mol.Biol., 371, 2007
|
|
2G8Y
 
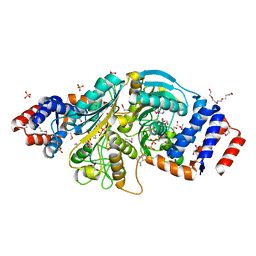 | | The structure of a putative malate/lactate dehydrogenase from E. coli. | | Descriptor: | 1,2-ETHANEDIOL, Malate/L-lactate dehydrogenases, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of a putative malate/lactate dehydrogenase from E. coli.
To be Published
|
|
2OOZ
 
 | | Macrophage Migration Inhibitory Factor (MIF) Complexed with OXIM6 (an OXIM Derivative Not Containing a Ring in its R-group) | | Descriptor: | 4-HYDROXYBENZALDEHYDE O-(3,3-DIMETHYLBUTANOYL)OXIME, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
2LAL
 
 | | CRYSTAL STRUCTURE DETERMINATION AND REFINEMENT AT 2.3 ANGSTROMS RESOLUTION OF THE LENTIL LECTIN | | Descriptor: | CALCIUM ION, LENTIL LECTIN (ALPHA CHAIN), LENTIL LECTIN (BETA CHAIN), ... | | Authors: | Loris, R, Steyaert, J, Maes, D, Lisgarten, J, Pickersgill, R, Wyns, L. | | Deposit date: | 1993-06-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of two crystal forms of lentil lectin at 1.8 A resolution.
Proteins, 20, 1994
|
|
3NZS
 
 | |
3WD4
 
 | | Serratia marcescens Chitinase B complexed with azide inhibitor and quinoline compound | | Descriptor: | (E)-N-(prop-2-en-1-yloxy)-1-(quinolin-4-yl)methanimine, Chitinase B, GLYCEROL, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6M32
 
 | | Cryo-EM structure of FMO-RC complex from green sulfur bacteria | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Chen, J.H, Zhang, X. | | Deposit date: | 2020-03-02 | | Release date: | 2020-11-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the photosynthetic complex from a green sulfur bacterium.
Science, 370, 2020
|
|
1XKF
 
 | |
2G6G
 
 | | Crystal structure of MltA from Neisseria gonorrhoeae | | Descriptor: | GLYCEROL, GNA33, SULFATE ION | | Authors: | Powell, A.J, Liu, Z.J, Nicholas, R.A, Davies, C. | | Deposit date: | 2006-02-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of the Lytic Transglycosylase MltA from N.gonorrhoeae and E.coli: Insights into Interdomain Movements and Substrate Binding.
J.Mol.Biol., 359, 2006
|
|
2OV4
 
 | |
2OWX
 
 | | THERMUS THERMOPHILUS AMYLOMALTASE AT pH 5.6 | | Descriptor: | 4-alpha-glucanotransferase, GLYCEROL, MALONATE ION | | Authors: | Barends, T.R.M, Kaper, T, Bultema, J.J, Dijkhuizen, L, van der Maarel, J.E.C, Dijkstra, B.W. | | Deposit date: | 2007-02-17 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-way stabilization of the covalent intermediate in amylomaltase, an alpha-amylase-like transglycosylase.
J.Biol.Chem., 282, 2007
|
|
4C1P
 
 | | Geobacillus thermoglucosidasius GH family 52 xylosidase | | Descriptor: | BETA-XYLOSIDASE, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Espina, G, Eley, K, Schneider, T.R, Crennell, S.J, Danson, M.J. | | Deposit date: | 2013-08-13 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | A Novel Beta-Xylosidase Structure from Geobacillus Thermoglucosidasius: The First Crystal Structure of a Glycoside Hydrolase Family Gh52 Enzyme Reveals Unpredicted Similarity to Other Glycoside Hydrolase Folds
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C2N
 
 | | Crystal structure of human testis angiotensin-I converting enzyme mutant E403R | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ANGIOTENSIN-CONVERTING ENZYME, CHLORIDE ION, ... | | Authors: | Masuyer, G, Yates, C.J, Schwager, S.L.U, Mohd, A, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2013-08-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular and Thermodynamic Mechanisms of the Chloride Dependent Human Angiotensin-I Converting Enzyme (Ace)
J.Biol.Chem., 289, 2014
|
|
2B45
 
 | | Crystal structure of an engineered uninhibited Bacillus subtilis xylanase in free state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endo-1,4-beta-xylanase A | | Authors: | Sansen, S, Dewilde, M, De Ranter, C.J, Sorensen, J.F, Sibbesen, O, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Triticum xylanse inhibitor-I in complex with bacillus subtilis xylanase
To be Published
|
|
4K5Z
 
 | | Crystal Structure of Human Chymase in Complex with Fragment Inhibitor 6-chloro-2,3-dihydro-1H-isoindol-1-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-2,3-dihydro-1H-isoindol-1-one, Chymase, ... | | Authors: | Collins, B.K, Padyana, A.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent, Selective Chymase Inhibitors via Fragment Linking Strategies.
J.Med.Chem., 56, 2013
|
|
3ABX
 
 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with p-nitrophenyl beta-D-cellotrioside | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, Cellobiohydrolase, MAGNESIUM ION | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-12-24 | | Release date: | 2010-01-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3ABZ
 
 | | Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus | | Descriptor: | Beta-glucosidase I, GLYCEROL | | Authors: | Yoshida, E, Hidaka, M, Fushinobu, S, Katayama, T, Kumagai, H. | | Deposit date: | 2009-12-25 | | Release date: | 2010-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus.
Biochem.J., 431, 2010
|
|
5POF
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10941a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, N-(1-benzylpiperidin-4-yl)acetamide, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
1QZR
 
 | | CRYSTAL STRUCTURE OF THE ATPASE REGION OF SACCHAROMYCES CEREVISIAE TOPOISOMERASE II BOUND TO ICRF-187 (DEXRAZOXANE) | | Descriptor: | (S)-4,4'-(1-METHYL-1,2-ETHANEDIYL)BIS-2,6-PIPERAZINEDIONE, DNA topoisomerase II, MAGNESIUM ION, ... | | Authors: | Classen, S, Olland, S, Berger, J.M. | | Deposit date: | 2003-09-17 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4KDP
 
 | | TcaR-ssDNA complex crystal structure reveals the novel ssDNA binding mechanism of the MarR family proteins | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(*CP*GP*CP*AP*GP*CP*GP*CP*GP*CP*AP*GP*CP*CP*CP*TP*A)-3'), ... | | Authors: | Chang, Y.M, Chen, C.K.-M, Wang, A.H.-J. | | Deposit date: | 2013-04-25 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | TcaR-ssDNA complex crystal structure reveals new DNA binding mechanism of the MarR family proteins.
Nucleic Acids Res., 42, 2014
|
|
3RVZ
 
 | | Crystal structure of the NavAb voltage-gated sodium channel (Ile217Cys, 2.8 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
2PVR
 
 | | Crystal structure of the catalytic subunit of protein kinase CK2 (C-terminal deletion mutant 1-335) in complex with two sulfate ions | | Descriptor: | Casein kinase II subunit alpha, catalytic subunit, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Niefind, K, Yde, C.W, Ermakova, I, Issinger, O.-G. | | Deposit date: | 2007-05-10 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Evolved to Be Active: Sulfate Ions Define Substrate Recognition Sites of CK2alpha and Emphasise its Exceptional Role within the CMGC Family of Eukaryotic Protein Kinases
J.Mol.Biol., 370, 2007
|
|
