8OIJ
 
 | | Drosophila Smaug-Smoothened complex | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Protein Smaug, ... | | Authors: | Ubartaite, G, Kubikova, J, Jeske, M. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for binding of Drosophila Smaug to the GPCR Smoothened and to the germline inducer Oskar.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8OFN
 
 | | Structure of the yellow fever virus (Asibi strain) dimeric envelope protein | | Descriptor: | Envelope glycoprotein, SULFATE ION | | Authors: | Covernton, E, Vaney, M.C, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | New insight into flavivirus maturation from structure/function studies of the yellow fever virus envelope protein complex.
Mbio, 14, 2023
|
|
6WTB
 
 | | Sort-Tagged Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Schneps, C.M, Crane, B.R. | | Deposit date: | 2020-05-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Tuning flavin environment to detect and control light-induced conformational switching in Drosophila cryptochrome.
Commun Biol, 4, 2021
|
|
6X38
 
 | |
7EDR
 
 | |
7ME4
 
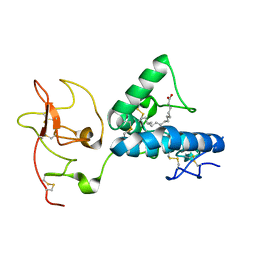 | | Structure of the extracellular WNT-binding module in Drosophila Ror2/Nrk | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITOLEIC ACID, Tyrosine-protein kinase transmembrane receptor Ror2 | | Authors: | Mendrola, J.M, Shi, F, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
7ME5
 
 | | Structure of the extracellular WNT-binding module in Drl-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tyrosine-protein kinase transmembrane receptor DRL-2 | | Authors: | Shi, F, Mendrola, J.M, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
7MSJ
 
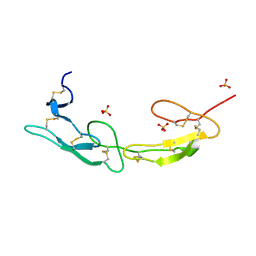 | | The crystal structure of mouse HVEM | | Descriptor: | SULFATE ION, Tumor necrosis factor receptor superfamily member 14 | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7MSG
 
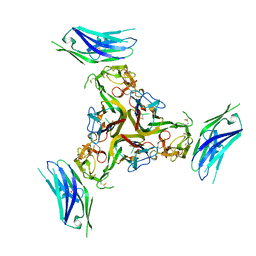 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
6XWS
 
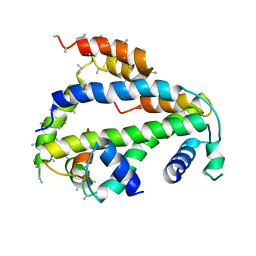 | | Crystal Structure of Drosophila CAL1 1-160 bound to CENP-A/H4 | | Descriptor: | Chromosome alignment defect 1,Chromosome alignment defect 1, Histone H3-like centromeric protein cid, Histone H4 | | Authors: | Jeyaprakash, A.A, Medina-Pritchard, B, Lazou, V, Zou, J, Byron, O, Abad, M.A, Rappsilber, J, Heun, P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.36 Å) | | Cite: | Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1.
Embo J., 39, 2020
|
|
7LCY
 
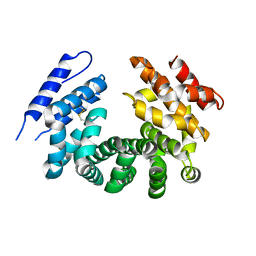 | | Crystal structure of the ligand-free ARM domain from Drosophila SARM1 | | Descriptor: | Isoform B of NAD(+) hydrolase sarm1 | | Authors: | Gu, W, Nanson, J.D, Luo, Z, McGuinness, H.Y, Manik, M.K, Jia, X, Ve, T, Kobe, B. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7LCZ
 
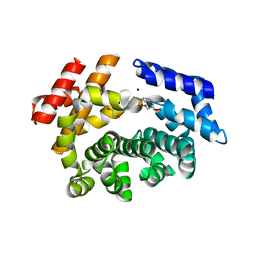 | | Crystal structure of the ARM domain from Drosophila SARM1 in complex with NMN | | Descriptor: | 1,2-ETHANEDIOL, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Isoform B of NAD(+) hydrolase sarm1, ... | | Authors: | Gu, W, Nanson, J.D, Luo, Z, Jia, X, Manik, M.K, Ve, T, Kobe, B. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7M6K
 
 | |
7MKK
 
 | |
7EXY
 
 | |
6O9L
 
 | | Human holo-PIC in the closed state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8PP7
 
 | | human RYBP-PRC1 bound to mononucleosome | | Descriptor: | DNA (215-mer), E3 ubiquitin-protein ligase RING2, Histone H2A, ... | | Authors: | Ciapponi, M, Benda, C, Mueller, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1.
Nat.Struct.Mol.Biol., 2024
|
|
8Q52
 
 | | A PBP-like protein built from fragments of different folds | | Descriptor: | Leucine-specific-binding protein,Chemotaxis protein CheY, SULFATE ION | | Authors: | Shanmugaratnam, S, Toledo-Patino, S, Goetz, S.K, Farias-Rico, J.A, Hocker, B. | | Deposit date: | 2023-08-08 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular handcraft of a well-folded protein chimera.
Febs Lett., 598, 2024
|
|
6OZ9
 
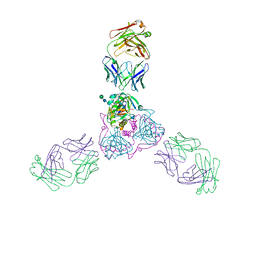 | | Ebola virus glycoprotein in complex with EBOV-520 Fab | | Descriptor: | EBOV-520 Fab heavy chain, EBOV-520 Fab light chain, Envelope glycoprotein, ... | | Authors: | Milligan, J.C, Altman, P.X, Hui, S, Hastie, K.M, Gilchuk, P, Crowe, J.E, Saphire, E.O. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.462 Å) | | Cite: | Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Immunity, 52, 2020
|
|
6P50
 
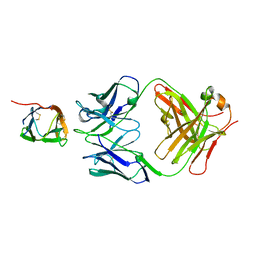 | | Crystal Structure of a Complex of human IL-7Ralpha with an anti-IL-7Ralpha Fab 4A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-7 receptor subunit alpha, anti-IL-7R 4A10 Fab heavy chain, ... | | Authors: | Walsh, S.T.R, Kashi, L, Kohnhorst, C.L. | | Deposit date: | 2019-05-29 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | New anti-IL-7R alpha monoclonal antibodies show efficacy against T cell acute lymphoblastic leukemia in pre-clinical models.
Leukemia, 34, 2020
|
|
6PJU
 
 | |
8S9H
 
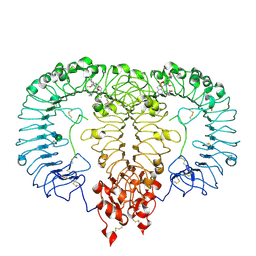 | | Crystal structure of monkey TLR7 ectodomain with compound 14 | | Descriptor: | (3S)-3-{[5-amino-1-({3-methoxy-5-[1-(oxan-4-yl)piperidin-4-yl]pyridin-2-yl}methyl)-1H-pyrazolo[4,3-d]pyrimidin-7-yl]amino}hexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Critton, D.A. | | Deposit date: | 2023-03-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.437 Å) | | Cite: | Identification and Optimization of Small Molecule Pyrazolopyrimidine TLR7 Agonists for Applications in Immuno-oncology.
Acs Med.Chem.Lett., 15, 2024
|
|
6P69
 
 | |
6PJR
 
 | |
6P91
 
 | | Structure of Lassa virus glycoprotein bound to Fab 18.5C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 18.5C Antibody heavy chain, ... | | Authors: | Saphire, E.O, Hastie, K.M. | | Deposit date: | 2019-06-09 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Convergent Structures Illuminate Features for Germline Antibody Binding and Pan-Lassa Virus Neutralization.
Cell, 178, 2019
|
|
