6USO
 
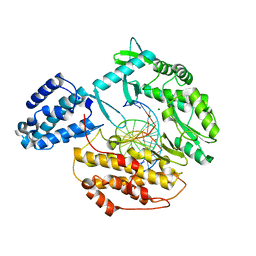 | |
1YG4
 
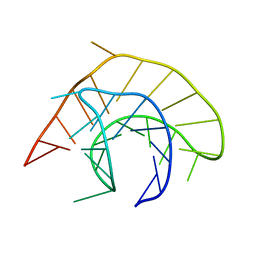 | |
6SYF
 
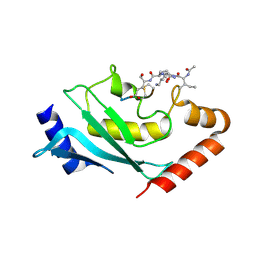 | | Human Ubc9 with covalent isopeptide ligand | | Descriptor: | ACE-ILE-LYS-GLN-GLU, ACE-LEU-ARG-LEU-ARG-GLY-CYS, SUMO-conjugating enzyme UBC9 | | Authors: | Hofmann, R, Akimoto, G, Wucherpfennig, T.G, Zeymer, C, Bode, J.W. | | Deposit date: | 2019-09-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lysine acylation using conjugating enzymes for site-specific modification and ubiquitination of recombinant proteins.
Nat.Chem., 12, 2020
|
|
1HRH
 
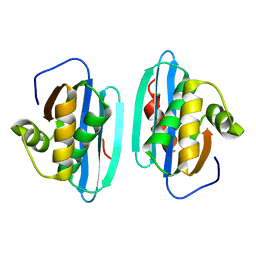 | |
4MK2
 
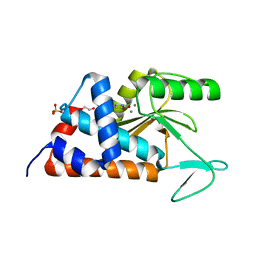 | | 3-(5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl)benzonitrile bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 1,2-ETHANEDIOL, 3-(5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl)benzonitrile, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
7ZNT
 
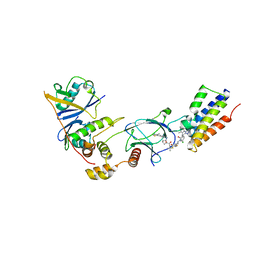 | | CRYSTAL STRUCTURE OF AT7 IN COMPLEX WITH THE SECOND BROMODOMAIN OF HUMAN BRD4 AND PVHL:ELONGINC:ELONGINB | | Descriptor: | (2~{S},4~{R})-1-[(2~{R})-3-[6-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]hexylsulfanyl]-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Elongin-B, ... | | Authors: | Hughes, S.J, Casement, R, Ciulli, A. | | Deposit date: | 2022-04-22 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional E3 ligase hotspots and resistance mechanisms to small-molecule degraders.
Nat.Chem.Biol., 19, 2023
|
|
3KQL
 
 | |
4MK1
 
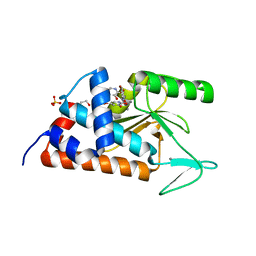 | | 5-bromopyridine-2,3-diol bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-3-hydroxypyridin-2(1H)-one, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors.
Acs Chem.Biol., 8, 2013
|
|
3KQN
 
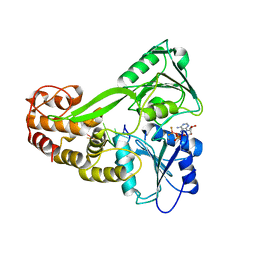 | |
3KQU
 
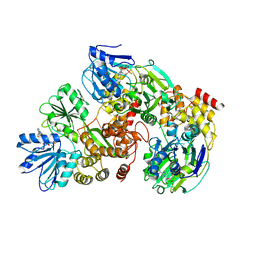 | |
4ZF7
 
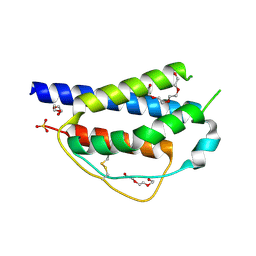 | | Crystal structure of ferret interleukin-2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Interleukin 2, PENTAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Newman, J, McKinstry, W.J, Adams, T.E. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-04 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structural and functional characterisation of ferret interleukin-2.
Dev.Comp.Immunol., 55, 2015
|
|
1MIV
 
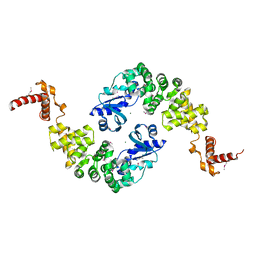 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme | | Descriptor: | MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIW
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
3LKZ
 
 | |
1MIY
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
8QXB
 
 | | TDP-43 amyloid fibrils: Morphology-2 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QXA
 
 | | TDP-43 amyloid fibrils: Morphology-1b | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QX9
 
 | | TDP-43 amyloid fibrils: Morphology-1a | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
5FPY
 
 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 5-bromo-1-methyl-1H-indole-2-carboxylic acid (AT21457) in an alternate binding site. | | Descriptor: | 5-bromo-1-methyl-1H-indole-2-carboxylic acid, SERINE PROTEASE NS3 | | Authors: | Davies, T.G, Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8PC2
 
 | | SelDeg51 in complex with FKBP51FK1 domain and pVHL:EloB:EloC | | Descriptor: | Elongin-B, Elongin-C, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Meyners, C, Walz, M, Geiger, T.M, Hausch, F. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Potent Proteolysis Targeting Chimera Enables Targeting the Scaffolding Functions of FK506-Binding Protein 51 (FKBP51).
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5FPT
 
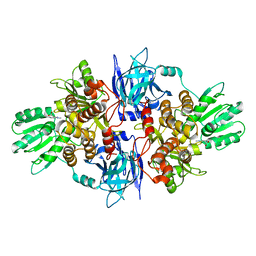 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 2-(1-methyl-1H-indol-3-yl)acetic acid (AT3437) in an alternate binding site. | | Descriptor: | (1-methyl-1H-indol-3-yl)acetic acid, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Pathuri, P, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8P5E
 
 | |
8P63
 
 | |
8P62
 
 | |
7K0W
 
 | |
