9CTP
 
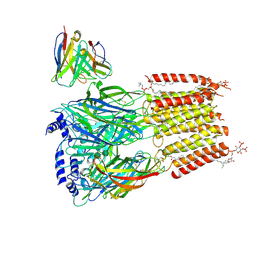 | | Native human GABAA receptor of beta2-alpha1-beta2-alpha3-gamma2 assembly | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, J, Hibbs, R.E, Noviello, C.M. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Resolving native GABA A receptor structures from the human brain.
Nature, 638, 2025
|
|
1R9Q
 
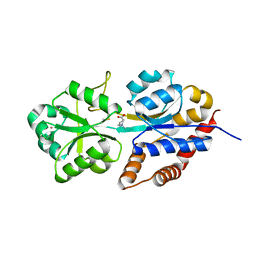 | | structure analysis of ProX in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding periplasmic protein, UNKNOWN ATOM OR ION | | Authors: | Schiefner, A, Breed, J, Bosser, L, Kneip, S, Gade, J, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cation-pi Interactions as Determinants for Binding of the Compatible Solutes Glycine Betaine and Proline Betaine by the Periplasmic Ligand-binding Protein ProX from Escherichia coli
J.BIOL.CHEM., 279, 2004
|
|
3QA3
 
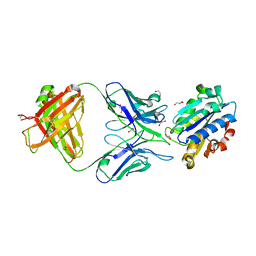 | | Crystal Structure of A-domain in complex with antibody | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy chain, Antibody Light chain, ... | | Authors: | Mahalingam, B, Xiong, J.P, Arnaout, M.A. | | Deposit date: | 2011-01-10 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stable Coordination of the Inhibitory Ca2+ Ion at the Metal Ion-Dependent Adhesion Site in Integrin CD11b/CD18 by an Antibody-Derived Ligand Aspartate: Implications for Integrin Regulation and Structure-Based Drug Design.
J.Immunol., 187, 2011
|
|
4B3K
 
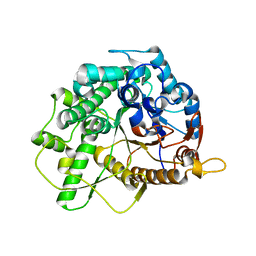 | | Family 1 6-phospho-beta-D glycosidase from Streptococcus pyogenes | | Descriptor: | BETA-GLUCOSIDASE | | Authors: | Stepper, J, Dabin, J, Ekloef, J.M, Thongpoo, P, Kongsaeree, P.T, Taylor, E.J, Turkenburg, J.P, Brumer, H, Davies, G.J. | | Deposit date: | 2012-07-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Activity of the Streptococcus Pyogenes Family Gh1 6-Phospho Beta-Glycosidase Spy1599
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FZ0
 
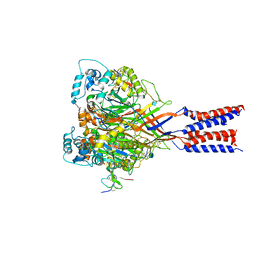 | |
7GFV
 
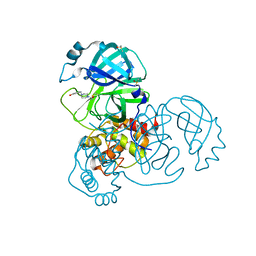 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-2db6411e-2 (Mpro-x11852) | | Descriptor: | 1-{4-[(3-chloro-5-hydroxyphenyl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4M6R
 
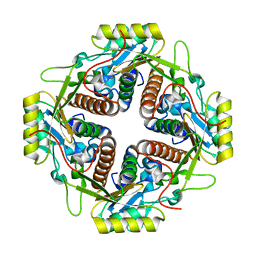 | | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme | | Descriptor: | Methylthioribulose-1-phosphate dehydratase, ZINC ION | | Authors: | Kang, W, Hong, S.H, Lee, H.M, Kim, N.Y, Lim, Y.C, Le, L.T.M, Lim, B, Kim, H.C, Kim, T.Y, Ashida, H, Yokota, A, Hah, S.S, Chun, K.H, Jung, Y.K, Yang, J.K. | | Deposit date: | 2013-08-10 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5P9J
 
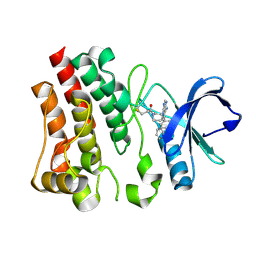 | | BTK1 COCRYSTALLIZED WITH IBRUTINIB | | Descriptor: | 1-[(3~{R})-3-[4-azanyl-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
5PAE
 
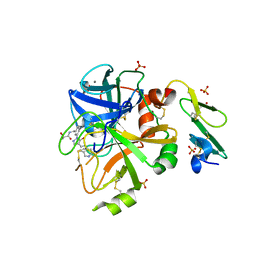 | | Crystal Structure of Factor VIIa in complex with (2S)-2-hydroxy-N-[[3-[5-hydroxy-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)pyrazol-1-yl]phenyl]methyl]propanamide | | Descriptor: | (2S)-2-hydroxy-N-[[3-[5-hydroxy-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)pyrazol-1-yl]phenyl]methyl]propanamide, CALCIUM ION, Coagulation factor VII heavy chain, ... | | Authors: | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | Deposit date: | 2016-11-10 | | Release date: | 2017-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a Factor VIIa complex
To be published
|
|
2ZPI
 
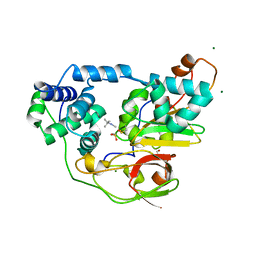 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 440min at 293K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2QG6
 
 | |
5PB6
 
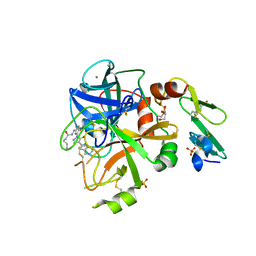 | | Crystal Structure of Factor VIIa in complex with N-[[3-[5-hydroxy-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)pyrazol-1-yl]phenyl]methyl]pentanamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Coagulation factor VII heavy chain, ... | | Authors: | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | Deposit date: | 2016-11-16 | | Release date: | 2017-06-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Factor VIIa complex
To be published
|
|
2GC8
 
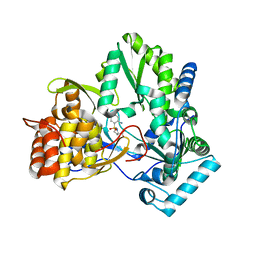 | | Structure of a Proline Sulfonamide Inhibitor Bound to HCV NS5b Polymerase | | Descriptor: | 1-[(2-AMINO-4-CHLORO-5-METHYLPHENYL)SULFONYL]-L-PROLINE, RNA-directed RNA polymerase | | Authors: | Gopalsamy, A, Chopra, R, Lim, K, Ciszewski, G, Shi, M, Curran, K.J, Sukits, S.F, Svenson, K, Bard, J, Ellingboe, J.W, Agarwal, A, Krishnamurthy, G, Howe, A.Y, Orlowski, M, Feld, B, O'connell, J, Mansour, T.S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Proline Sulfonamides as Potent and Selective Hepatitis C Virus NS5b Polymerase Inhibitors. Evidence for a New NS5b Polymerase Binding Site.
J.Med.Chem., 49, 2006
|
|
6LE7
 
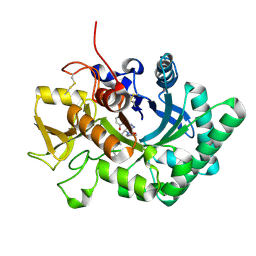 | | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2 | | Descriptor: | (4R)-3-(2-hydroxyphenyl)-4-(3-methoxy-4-propoxy-phenyl)-5-(pyridin-3-ylmethyl)-1,4-dihydropyrrolo[3,4-c]pyrazol-6-one, Probable endochitinase | | Authors: | Chen, Q, Yang, Q, Zhou, Y. | | Deposit date: | 2019-11-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85753441 Å) | | Cite: | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2
To Be Published
|
|
3RII
 
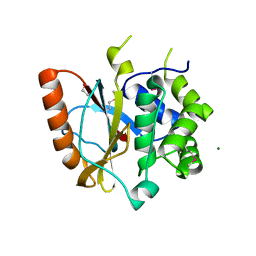 | | Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Das, C. | | Deposit date: | 2011-04-13 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.0008 Å) | | Cite: | Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme.
Febs J., 278, 2011
|
|
4M0Y
 
 | |
7GDN
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-0e996074-1 (Mpro-x11159) | | Descriptor: | (4R)-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4AZC
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | (2S,3aR,5R,6S,7R,7aR)-5-(hydroxymethyl)-2-methyl-2,3a,5,6,7,7a-hexahydro-1H-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
3NAY
 
 | | PDK1 in complex with inhibitor MP6 | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 4-(2-cyclopropylethylidene)-9-(1H-pyrazol-4-yl)-6-{[(1R)-1,2,2-trimethylpropyl]amino}benzo[c][1,6]naphthyridin-1(4H)-one | | Authors: | Yan, Y, Munshi, S.K, Allison, T. | | Deposit date: | 2010-06-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective inhibition of PDK1 using an allosteric kinase inhibitor and RNAi impairs cancer cell migration and anchorage-independent growth of primary tumor lines
J.Biol.Chem., 2010
|
|
7GHA
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAN-LON-a5fc619e-8 (Mpro-x3077) | | Descriptor: | 1-{(1S,4S)-5-[(3-chlorophenyl)methyl]-2,5-diazabicyclo[2.2.1]heptan-2-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5POW
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10894b | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, N-[(pyridin-2-yl)methyl]acetamide, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
7GM7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-a577c8a2-1 (Mpro-P2099) | | Descriptor: | (4S)-6-chloro-N-(isoquinolin-4-yl)-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5PP7
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 7) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PQE
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 51) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
1URR
 
 | |
