1Z5X
 
 | | hemipteran ecdysone receptor ligand-binding domain complexed with ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor ligand binding domain, PHOSPHATE ION, ... | | Authors: | Carmichael, J.A, Lawrence, M.C, Graham, L.D, Pilling, P.A, Epa, V.C, Noyce, L, Lovrecz, G, Winkler, D.A, Pawlak-Skrzecz, A. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | The X-ray structure of a hemipteran ecdysone receptor ligand-binding domain: comparison with a lepidopteran ecdysone receptor ligand-binding domain and implications for insecticide design.
J.Biol.Chem., 280, 2005
|
|
7W0Z
 
 | | Glycosyltranferase UGT74AN2 | | Descriptor: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7ZYS
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2227 | | Descriptor: | 1-[(8~{R},15~{S},18~{S})-18-(4-azanylbutyl)-15-butyl-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(26),22,24-trien-8-yl]guanidine, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-05-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.
Arch Pharm, 2024
|
|
1Z23
 
 | | The serine-rich domain from Crk-associated substrate (p130Cas) | | Descriptor: | CRK-associated substrate | | Authors: | Briknarova, K, Nasertorabi, F, Havert, M.L, Eggleston, E, Hoyt, D.W, Li, C, Olson, A.J, Vuori, K, Ely, K.R. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The serine-rich domain from Crk-associated substrate (p130cas) is a four-helix bundle.
J.Biol.Chem., 280, 2005
|
|
4I49
 
 | | Structure of ngNAGS bound with bisubstrate analog CoA-NAG | | Descriptor: | (2S)-2-({(3S,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14,20-trioxo-2,4,6-trioxa-18-thia-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaicosan-20-yl}amino)pentanedioic acid (non-preferred name), Amino-acid acetyltransferase, SULFATE ION | | Authors: | Shi, D, Zhao, G, Allewell, N.M, Tuchman, M. | | Deposit date: | 2012-11-27 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the complex of Neisseria gonorrhoeae N-acetyl-l-glutamate synthase with a bound bisubstrate analog.
Biochem.Biophys.Res.Commun., 430, 2013
|
|
7ZYV
 
 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.13 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, ... | | Authors: | Sarewicz, M, Szwalec, M, Pintscher, S, Indyka, P, Rawski, M, Pietras, R, Mielecki, B, Koziej, L, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
4I32
 
 | | Crystal structure of HCV NS3/4A D168V protease complexed with compound 4 | | Descriptor: | (2R,6S,7E,10E,13aR,14aR,16aS)-2-{[7-methoxy-8-methyl-2-(propan-2-yloxy)quinolin-4-yl]oxy}-N-[(1-methylcyclopropyl)sulfonyl]-6-{[(1-methyl-1H-pyrazol-3-yl)carbonyl]amino}-5,16-dioxo-1,2,3,6,9,12,13,13a,14,15,16,16a-dodecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamide, Genome polyprotein, HCV non-structural protein 4A, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-02 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.3001 Å) | | Cite: | Molecular Mechanism by Which a Potent Hepatitis C Virus NS3-NS4A Protease Inhibitor Overcomes Emergence of Resistance.
J.Biol.Chem., 288, 2013
|
|
2A4Q
 
 | | HCV NS3 protease with NS4a peptide and a covalently bound macrocyclic ketoamide compound. | | Descriptor: | (2R)-({N-[(3S)-3-({[(3S,6S)-6-CYCLOHEXYL-5,8-DIOXO-4,7-DIAZABICYCLO[14.3.1]ICOSA-1(20),16,18-TRIEN-3-YL]CARBONYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETIC ACID, BETA-MERCAPTOETHANOL, NS3 protease/helicase', ... | | Authors: | Chen, K.X, Njoroge, F.G, Prongay, A, Pichardo, J, Madison, V, Girijavallabhan, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and Biological Activity of Macrocyclic Inhibitors of Hepatitis C Virus (HCV) NS3 Protease
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4IJ7
 
 | | Crystal structure of Odorant Binding Protein 48 from Anopheles gambiae (AgamOBP48) with PEG | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Odorant binding protein-8, SODIUM ION | | Authors: | Zographos, S.E, Tsitsanou, K.E, Drakou, C.E. | | Deposit date: | 2012-12-21 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal and Solution Studies of the "Plus-C" Odorant-binding Protein 48 from Anopheles gambiae: CONTROL OF BINDING SPECIFICITY THROUGH THREE-DIMENSIONAL DOMAIN SWAPPING.
J.Biol.Chem., 288, 2013
|
|
8A15
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2230 | | Descriptor: | 1-[(8~{R},15~{S},18~{S})-15-(4-azanylbutyl)-18-(naphthalen-2-ylmethyl)-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-05-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.
Arch Pharm, 2024
|
|
2AZC
 
 | | HIV-1 Protease NL4-3 6X mutant | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-10 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
2E9V
 
 | | Structure of h-CHK1 complexed with A859017 | | Descriptor: | 18-CHLORO-2-OXO-17-[(PYRIDIN-4-YLMETHYL)AMINO]-2,3,11,12,13,14-HEXAHYDRO-1H,10H-4,8-(AZENO)-9,15,1,3,6-BENZODIOXATRIAZACYCLOHEPTADECINE-7-CARBONITRILE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-27 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis and Biological Evaluation of Potent Selective Macrocyclic Chk1 Inhibitors
To be Published
|
|
2E9U
 
 | | Structure of h-CHK1 complexed with A780125 | | Descriptor: | 18-CHLORO-11,12,13,14-TETRAHYDRO-1H,10H-8,4-(AZENO)-9,15,1,3,6-BENZODIOXATRIAZACYCLOHEPTADECIN-2-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-27 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis and Biological Evaluation of Potent Selective Macrocyclic Chk1 Inhibitors
To be Published
|
|
2AZB
 
 | | HIV-1 Protease NL4-3 3X mutant in complex with inhibitor, TL-3 | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
2E27
 
 | | Crystal structure of Fv fragment of anti-ciguatoxin antibody complexed with ABC-ring of ciguatoxin | | Descriptor: | 3,7:6,10:9,14-TRIANHYDRO-2,5,11,12,13-PENTADEOXY-4-O-(METHOXYMETHYL)-L-ARABINO-L-ALLO-TETRADEC-12-ENONIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Yokota, A, Tanaka, Y, Tsumoto, K. | | Deposit date: | 2006-11-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Critical contribution of aromatic rings to specific recognition of polyether rings. The case of ciguatoxin CTX3C-ABC and its specific antibody 1C49.
J.Biol.Chem., 283, 2008
|
|
1RR8
 
 | | Structural Mechanisms of Camptothecin Resistance by Mutations in Human Topoisomerase I | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, 2-(1-DIMETHYLAMINOMETHYL-2-HYDROXY-8-HYDROXYMETHYL-9-OXO-9,11-DIHYDRO-INDOLIZINO[1,2-B]QUINOLIN-7-YL)-2-HYDROXY-BUTYRIC ACID, 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T*GP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', ... | | Authors: | Chrencik, J.E, Staker, B.L, Burgin, A.B, Stewart, L, Redinbo, M.R. | | Deposit date: | 2003-12-08 | | Release date: | 2004-07-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanisms of camptothecin resistance by human topoisomerase I mutations
J.Mol.Biol., 339, 2004
|
|
2AZ9
 
 | | HIV-1 Protease NL4-3 1X mutant | | Descriptor: | PROTEASE RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Kutilek, V, Morris, G.M, Lin, Y.-C, Elder, J.H, Torbett, B.E, Stout, C.D. | | Deposit date: | 2005-09-09 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Mechanisms of Drug Resistance in HIV-1 Protease NL4-3
J.Mol.Biol., 356, 2006
|
|
1QJX
 
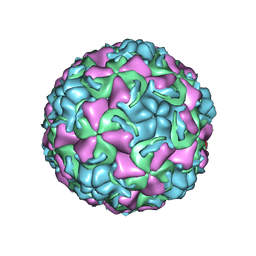 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND WIN68934 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[4-METHYL-2H-TETRAZOL-2-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QHJ
 
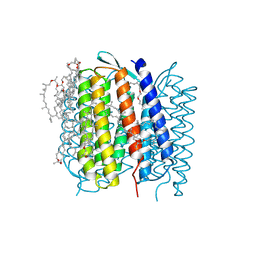 | | X-RAY STRUCTURE OF BACTERIORHODOPSIN GROWN IN LIPIDIC CUBIC PHASES | | Descriptor: | 1,2-[DI-2,6,10,14-TETRAMETHYL-HEXADECAN-16-OXY]-PROPANE, PROTEIN (BACTERIORHODOPSIN), RETINAL | | Authors: | Belrhali, H, Nollert, P, Royant, A, Menzel, C, Rosenbusch, J.P, Landau, E.M, Pebay-Peyroula, E. | | Deposit date: | 1999-05-04 | | Release date: | 1999-07-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 A resolution.
Structure Fold.Des., 7, 1999
|
|
1QJU
 
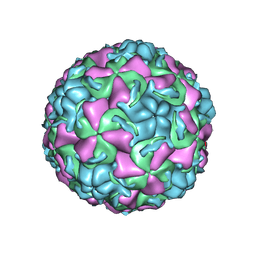 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP61209 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2N-METHYL-2H-TETRAZOL-5-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Minor, I, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-05 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QJY
 
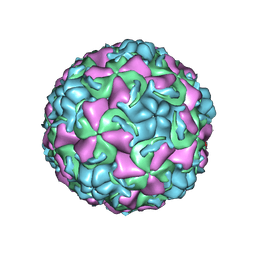 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP65099 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2-METHYL-4-ISOXAZOLYL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7ALR
 
 | | Crystal structure of TD1-gatorbulin1 complex | | Descriptor: | (2~{R})-2-oxidanyl-2-[(6~{S},9~{S},12~{S},15~{S},17~{S})-6,10,12,17-tetramethyl-3-methylidene-7-oxidanyl-2,5,8,11,14-pentakis(oxidanylidene)-13-oxa-1,4,7,10-tetrazabicyclo[13.3.0]octadecan-9-yl]ethanamide, Designed Ankyrin Repeat Protein (DARPIN) D1, GLYCEROL, ... | | Authors: | Oliva, M.A, Diaz, J.F. | | Deposit date: | 2020-10-07 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Gatorbulin-1, a distinct cyclodepsipeptide chemotype, targets a seventh tubulin pharmacological site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7X81
 
 | | The crystal structure of PloI4-C16M/D46A/I137V in complex with exo-2+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,11E,12aR,14aS,17E,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-11,12a,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,12a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)cyclobuta[b]naphtho[2,1-j][1]azacyclotetradecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-10 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
7X86
 
 | | The crystal structure of PloI4-F124L in complex with endo-4+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,10aS,13S,14aS,18aR,18bR,E)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,10a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)benzo[b]naphtho[2,1-h][1]azacyclododecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-11 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
2C78
 
 | | EF-Tu complexed with a GTP analog and the antibiotic pulvomycin | | Descriptor: | (1S,2S,3E,5E,7E,10S,11S,12S)-12-[(2R,4E,6E,8Z,10R,12E,14E,16Z,18S,19Z)-10,18-DIHYDROXY-12,16,19-TRIMETHYL-11,22-DIOXOOX ACYCLODOCOSA-4,6,8,12,14,16,19-HEPTAEN-2-YL]-2,11-DIHYDROXY-1,10-DIMETHYL-9-OXOTRIDECA-3,5,7-TRIEN-1-YL 6-DEOXY-2,4-DI-O-METHYL-BETA-L-GALACTOPYRANOSIDE, ELONGATION FACTOR TU-A, MAGNESIUM ION, ... | | Authors: | Parmeggiani, A, Krab, I.M, Okamura, S, Nielsen, R.C, Nyborg, J, Nissen, P. | | Deposit date: | 2005-11-18 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu.
Biochemistry, 45, 2006
|
|
