2Y2P
 
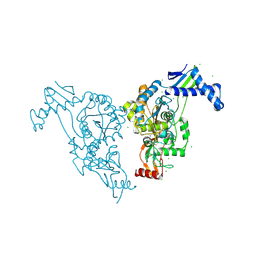 | | Penicillin-binding protein 1b (pbp-1b) in complex with an alkyl boronate (z10) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
5AYI
 
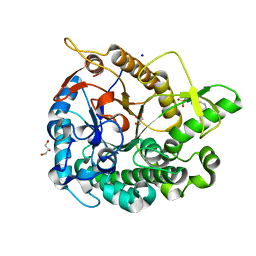 | | Crystal structure of GH1 Beta-glucosidase TD2F2 N223Q mutant | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, BETA-GLUCOSIDASE, GLYCEROL, ... | | Authors: | Jo, T, Manninen, J.A, Matsuzawa, T, Uchiyama, T, Yaoi, K, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-08-21 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and identification of a key amino acid for glucose tolerance, substrate specificity, and transglycosylation activity of metagenomic beta-glucosidase Td2F2
Febs J., 283, 2016
|
|
4ZXI
 
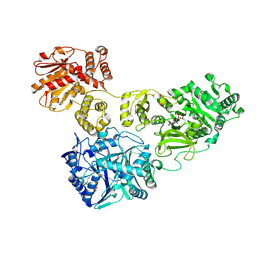 | | Crystal Structure of holo-AB3403 a four domain nonribosomal peptide synthetase bound to AMP and Glycine | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Drake, E.J, Miller, B.R, Allen, C.L, Gulick, A.M. | | Deposit date: | 2015-05-20 | | Release date: | 2015-12-30 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of two distinct conformations of holo-non-ribosomal peptide synthetases.
Nature, 529, 2016
|
|
7GH1
 
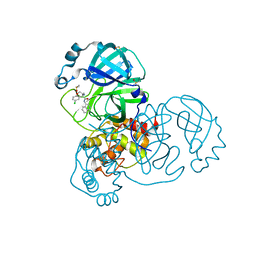 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-2bb0cf2b-1 (Mpro-x12731) | | Descriptor: | (4S)-4-(aminomethyl)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4ZYQ
 
 | |
2Y3A
 
 | | Crystal structure of p110beta in complex with icSH2 of p85beta and the drug GDC-0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT BETA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT BETA ISOFORM | | Authors: | Zhang, X, Vadas, O, Perisic, O, Williams, R.L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Lipid Kinase P110Beta-P85Beta Elucidates an Unusual Sh2-Domain-Mediated Inhibitory Mechanism.
Mol.Cell, 41, 2011
|
|
6M65
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with GMPPNP (GDP) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7GEU
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-50fe53e8-1 (Mpro-x11508) | | Descriptor: | 2-(3-chlorophenyl)-N-(phthalazin-1-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4QMZ
 
 | | MST3 IN COMPLEX WITH SUNITINIB | | Descriptor: | CHLORIDE ION, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, SERINE/THREONINE-PROTEIN KINASE 24 | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
7GFB
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-b3e365b9-1 (Mpro-x11612) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GDU
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with PET-SGC-a8a902d9-1 (Mpro-x11225) | | Descriptor: | (3R)-3-(4-hydroxypiperidin-1-yl)-N-(4-methylpyridin-3-yl)-3-(thiophen-3-yl)propanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
9KP2
 
 | | Crystal structure of Horse spleen L-ferritin mutant (E56F/R59F) with Nile Red | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-22 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
5B4A
 
 | | Crystal structure of LpxH with lipid X in spacegroup P21 | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
1YYO
 
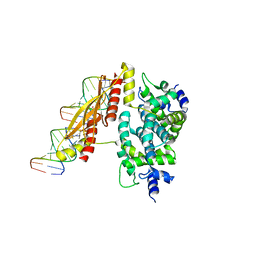 | | Crystal structure of RNase III mutant E110K from Aquifex aeolicus complexed with double-stranded RNA at 2.9-Angstrom Resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
3RZC
 
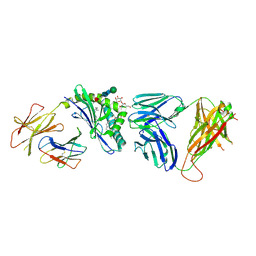 | | Structure of the self-antigen iGb3 bound to mouse CD1d and in complex with the iNKT TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, Beta-2-microglobulin, ... | | Authors: | Yu, E.D, Girardi, E, Wang, J, Zajonc, D.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cutting Edge: Structural Basis for the Recognition of {beta}-Linked Glycolipid Antigens by Invariant NKT Cells.
J.Immunol., 187, 2011
|
|
4Q4O
 
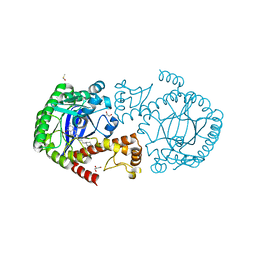 | | tRNA-Guanine Transglycosylase (TGT) in Complex with 6-Amino-2-{[2-(piperidin-1-yl)ethyl]amino}-1H,7H,8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-{[2-(piperidin-1-yl)ethyl]amino}-3,5-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Beyond Affinity: Enthalpy-Entropy Factorization Unravels Complexity of a Flat Structure-Activity Relationship for Inhibition of a tRNA-Modifying Enzyme.
J.Med.Chem., 57, 2014
|
|
9KP5
 
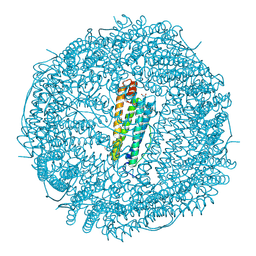 | | Crystal structure of Horse spleen L-ferritin mutant (E56F/R59F) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-22 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
5UVI
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
9KKR
 
 | | Crystal structure of Horse spleen L-ferritin mutant (E53F/E56F/E57F/R59F/E60F/E63F) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-14 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
2XV2
 
 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAHAAM), chemically reduced, pH4.2 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
1D2Z
 
 | | THREE-DIMENSIONAL STRUCTURE OF A COMPLEX BETWEEN THE DEATH DOMAINS OF PELLE AND TUBE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DEATH DOMAIN OF PELLE, DEATH DOMAIN OF TUBE | | Authors: | Xiao, T, Towb, P, Wasserman, S.A, Sprang, S.R. | | Deposit date: | 1999-09-28 | | Release date: | 1999-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a complex between the death domains of Pelle and Tube.
Cell(Cambridge,Mass.), 99, 1999
|
|
9KLE
 
 | | Crystal structure of Horse spleen L-ferritin mutant (R52F/E56F/R59F/E63F) with Nile Red | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-14 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
9KP7
 
 | | Crystal structure of Horse spleen L-ferritin mutant (R59F) with Nile Red | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, T, Hishikawa, Y, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-22 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of Aromatic Interaction Networks in a Protein Cage Modulated by Fluorescent Ligand Binding.
Adv Sci, 12, 2025
|
|
6M9U
 
 | | Structure of the apo-form of 20beta-Hydroxysteroid Dehydrogenase from Bifidobacterium adolescentis strain L2-32 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Mythen, S.M, Pollet, R.M, Koropatkin, N.M, Ridlon, J.M. | | Deposit date: | 2018-08-24 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of 20 beta-hydroxysteroid dehydrogenase fromBifidobacterium adolescentisstrain L2-32.
J.Biol.Chem., 294, 2019
|
|
4Q92
 
 | | 1.90 Angstrom resolution crystal structure of apo betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) with BME-modified Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
