1VY4
 
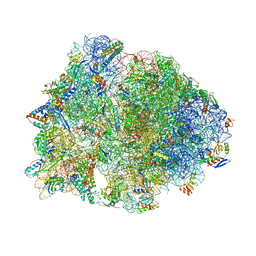 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing acylated tRNA-substrates in the A and P sites. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2J62
 
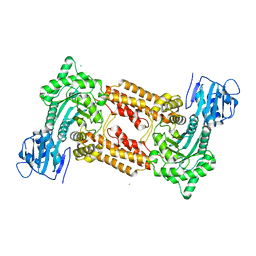 | | Structure of a bacterial O-glcnacase in complex with glcnacstatin | | Descriptor: | CHLORIDE ION, N-[(5R,6R,7R,8S)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-2-(2-PHENYLETHYL)-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDIN-8-YL]-2-METHYLPROPANAMIDE, O-GlcNAcase NagJ | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Schimpl, M, Shepherd, S.M, Shpiro, N.A, van Aalten, D.M.F. | | Deposit date: | 2006-09-22 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels.
J. Am. Chem. Soc., 128, 2006
|
|
1SJ4
 
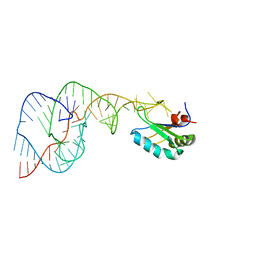 | | Crystal structure of a C75U mutant Hepatitis Delta Virus ribozyme precursor, in Cu2+ solution | | Descriptor: | precursor form of the Hepatitis Delta virus ribozyme, small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H, Doudna, J.A. | | Deposit date: | 2004-03-02 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme
catalysis
Nature, 429, 2004
|
|
3RKZ
 
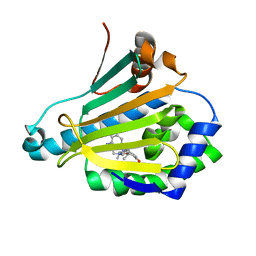 | | Discovery of a stable macrocyclic o-aminobenzamide Hsp90 inhibitor capable of significantly decreasing tumor volume in a mouse xenograft model. | | Descriptor: | (5R,6S)-3-(L-alanyl)-5,6,15,15,18-pentamethyl-17-oxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-12,8-(metheno)[1,5,9]triazacyclotetradecino[1,2-a]indole-9-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, Li, Z, Dushin, R.G, Nittoli, T, Otteng, M, Nikitenko, A, Golas, J.M, Liu, H, Lucas, J, Boschelli, F, Vogan, E, Olland, A, Johnson, M, Levin, J.I. | | Deposit date: | 2011-04-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5693 Å) | | Cite: | Discovery of a stable macrocyclic o-aminobenzamide Hsp90 inhibitor which significantly decreases tumor volume in a mouse xenograft model.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3GAR
 
 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
1Y8Y
 
 | | Crystal structure of human CDK2 complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | (5-CHLOROPYRAZOLO[1,5-A]PYRIMIDIN-7-YL)-(4-METHANESULFONYLPHENYL)AMINE, Cell division protein kinase 2 | | Authors: | Williamson, D.S, Parratt, M.J, Torrance, C.J, Bower, J.F, Moore, J.D, Richardson, C.M, Dokurno, P, Cansfield, A.D, Francis, G.L, Hebdon, R.J, Howes, R, Jackson, P.S, Lockie, A.M, Murray, J.B, Nunns, C.L, Powles, J, Robertson, A, Surgenor, A.E. | | Deposit date: | 2004-12-14 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structure-guided design of pyrazolo[1,5-a]pyrimidines as inhibitors of human cyclin-dependent kinase 2.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1Y91
 
 | | Crystal structure of human CDK2 complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | 4-[5-(TRANS-4-AMINOCYCLOHEXYLAMINO)-3-ISOPROPYLPYRAZOLO[1,5-A]PYRIMIDIN-7-YLAMINO]-N,N-DIMETHYLBENZENESULFONAMIDE, Cell division protein kinase 2 | | Authors: | Williamson, D.S, Parratt, M.J, Torrance, C.J, Bower, J.F, Moore, J.D, Richardson, C.M, Dokurno, P, Cansfield, A.D, Francis, G.L, Hebdon, R.J, Howes, R, Jackson, P.S, Lockie, A.M, Murray, J.B, Nunns, C.L, Powles, J, Robertson, A, Surgenor, A.E. | | Deposit date: | 2004-12-14 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-guided design of pyrazolo[1,5-a]pyrimidines as inhibitors of human cyclin-dependent kinase 2.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1I1A
 
 | | CRYSTAL STRUCTURE OF THE NEONATAL FC RECEPTOR COMPLEXED WITH A HETERODIMERIC FC | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Martin, W.L, West Jr, A.P, Gan, L, Bjorkman, P.J. | | Deposit date: | 2001-01-31 | | Release date: | 2001-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding.
Mol.Cell, 7, 2001
|
|
3NRM
 
 | | Imidazo[1,2-a]pyrazine-based Aurora Kinase Inhibitors | | Descriptor: | N-(3-methylisothiazol-5-yl)-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-amine, Serine/threonine-protein kinase 6 | | Authors: | Hruza, A. | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of imidazo[1,2-a]pyrazine-based Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1MEY
 
 | | CRYSTAL STRUCTURE OF A DESIGNED ZINC FINGER PROTEIN BOUND TO DNA | | Descriptor: | CHLORIDE ION, CONSENSUS ZINC FINGER, DNA (5'-D(*AP*TP*GP*AP*GP*GP*CP*AP*GP*AP*AP*CP*T)-3'), ... | | Authors: | Kim, C.A, Berg, J.M. | | Deposit date: | 1996-09-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A 2.2 A Resolution Crystal Structure of a Designed Zinc Finger Protein Bound to DNA
Nat.Struct.Biol., 3, 1996
|
|
3ZW3
 
 | | Fragment based discovery of a novel and selective PI3 Kinase inhibitor | | Descriptor: | N-{6-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]imidazo[1,2-a]pyridin-2-yl}acetamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Brown, D.G, Hughes, S.J, Milan, D.S, Kilty, I.C, Lewthwaite, R.A, Mathias, J.P, O'Reilly, M.A, Pannifer, A, Phelan, A, Baldock, D.A. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment based discovery of a novel and selective PI3 kinase inhibitor.
Bioorg. Med. Chem. Lett., 21, 2011
|
|
1IT6
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CALYCULIN A AND THE CATALYTIC SUBUNIT OF PROTEIN PHOSPHATASE 1 | | Descriptor: | CALYCULIN A, MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 1 GAMMA (PP1-GAMMA) CATALYTIC SUBUNIT | | Authors: | Kita, A, Matsunaga, S, Takai, A, Kataiwa, H, Wakimoto, T, Fusetani, N, Isobe, M, Miki, K. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex between calyculin A and the catalytic subunit of protein phosphatase 1.
Structure, 10, 2002
|
|
1INV
 
 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
1QR0
 
 | | CRYSTAL STRUCTURE OF THE 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP-COENZYME A COMPLEX | | Descriptor: | 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP, COENZYME A, MAGNESIUM ION | | Authors: | Reuter, K, Mofid, R.M, Marahiel, A.M, Ficner, R. | | Deposit date: | 1999-06-17 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4'-phosphopantetheinyl transferase superfamily.
EMBO J., 18, 1999
|
|
3SDY
 
 | |
2IL4
 
 | | Crystal structure of At1g77540-Coenzyme A Complex | | Descriptor: | COENZYME A, Protein At1g77540 | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
3QSK
 
 | | 5 Histidine Variant of the anti-RNase A VHH in Complex with RNAse A | | Descriptor: | Engineered 5 Histidine anti-RNase A Camelid VHH Antibody Domain Variant, Ribonuclease pancreatic | | Authors: | Murtaugh, M.L, Fanning, S.W, Sharma, T.M, Terry, A.M, Horn, J.R. | | Deposit date: | 2011-02-21 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A combinatorial histidine scanning library approach to engineer highly pH-dependent protein switches.
Protein Sci., 20, 2011
|
|
1I7V
 
 | | THE SOLUTION STRUCTURE OF A BAY REGION 1R-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 1R,2S,3R,4S-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*GP*GP*AP*CP*AP*(BZA)AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Li, Z, Tamura, P.J, Wilkinson, A.S, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 2001-03-10 | | Release date: | 2001-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Intercalation of the (1R,2S,3R,4S)-N6-[1-(1,2,3,4-tetrahydro-2,3,4-trihydroxybenz[a]anthracenyl)]-2'-deoxyadenosyl adduct in the N-ras codon 61 sequence: DNA sequence effects
Biochemistry, 40, 2001
|
|
4ID7
 
 | | ACK1 kinase in complex with the inhibitor cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Descriptor: | Activated CDC42 kinase 1, SULFATE ION, cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Authors: | Jin, M, Wang, J, Kleinberg, A, Kadalbajoo, M, Siu, K, Cooke, A, Bittner, M, Yao, Y, Thelemann, A, Ji, Q, Bhagwat, S, Crew, A.P, Pachter, J, Epstein, D, Mulvihill, M.J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, selective and orally bioavailable imidazo[1,5-a]pyrazine derived ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5UZ4
 
 | | The cryo-EM structure of YjeQ bound to the 30S subunit suggests a fidelity checkpoint function for this protein in ribosome assembly | | Descriptor: | 16S RIBOSOMAL RNA, 3'-O-(N-methylanthraniloyl)-beta:gamma-imidoguanosine-5'-triphosphate, 30S ribosomal protein S10, ... | | Authors: | Razi, A, Guarne, A, Ortega, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | The cryo-EM structure of YjeQ bound to the 30S subunit suggests a fidelity checkpoint function for this protein in ribosome assembly.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1PN4
 
 | | Crystal structure of 2-enoyl-CoA hydratase 2 domain of Candida tropicalis multifunctional enzyme type 2 complexed with (3R)-hydroxydecanoyl-CoA. | | Descriptor: | 1,2-ETHANEDIOL, 3R-HYDROXYDECANOYL-COENZYME A, Peroxisomal hydratase-dehydrogenase-epimerase | | Authors: | Koski, M.K, Haapalainen, A.M, Hiltunen, J.K, Glumoff, T. | | Deposit date: | 2003-06-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Two-domain Structure of One Subunit Explains Unique Features of Eukaryotic Hydratase 2.
J.Biol.Chem., 279, 2004
|
|
1MPL
 
 | | CRYSTAL STRUCTURE OF PHOSPHONATE-INHIBITED D-ALA-D-ALA PEPTIDASE REVEALS AN ANALOG OF A TETRAHEDRAL TRANSITION STATE | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, GLYCEROL, GLYCYL-L-A-AMINOPIMELYL-E-(D-2-AMINOETHYL)PHOSPHONATE | | Authors: | Silvaggi, N.R, Anderson, J.W, Brinsmade, S.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2002-09-12 | | Release date: | 2003-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The Crystal Structure of Phosphonate-Inhibited
d-Ala-d-Ala Peptidase Reveals an Analogue of a Tetrahedral Transition State.
Biochemistry, 42, 2003
|
|
4CAS
 
 | | Serial femtosecond crystallography structure of a photosynthetic reaction center | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Johansson, L.C, Arnlund, D, Katona, G, White, T.A, Barty, A, DePonte, D.P, Shoeman, R.L, Wickstrand, C, Sharma, A, Williams, G.J, Aquila, A, Bogan, M.J, Caleman, C, Davidsson, J, Doak, R.B, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Kassemeyer, S, Kirian, R.A, Kupitz, C, Liang, M, Lomb, L, Malmerberg, E, Martin, A.V, Messerschmidt, M, Nass, K, Redecke, L, Seibert, M.M, Sjohamn, J, Steinbrener, J, Stellato, F, Wang, D, Wahlgren, W.Y, Weierstall, U, Westenhoff, S, Zatsepin, N.A, Boutet, S, Spence, J.C.H, Schlichting, I, Chapman, H.N, Fromme, P, Neutze, R. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography.
Nat Commun, 4, 2013
|
|
1KBY
 
 | | Structure of Photosynthetic Reaction Center with bacteriochlorophyll-bacteriopheophytin heterodimer | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C, Goetsch, A, Allen, J.P. | | Deposit date: | 2001-11-07 | | Release date: | 2002-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of the heterodimer reaction center from Rhodobacter sphaeroides at 2.55 a resolution.
Photosynth.Res., 74, 2002
|
|
1KVH
 
 | | NCSi-gb-bulge-DNA complex induced formation of a DNA bulge structure by a molecular wedge ligand-post-activated neocarzinostatin chromophore | | Descriptor: | 5'-D(*CP*CP*CP*GP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*TP*TP*CP*GP*GP*G)-3', SPIRO[[7-METHOXY-5-METHYL-1,2-DIHYDRO-NAPHTHALENE]-3,1'-[5-HYDROXY-9-[2-METHYLAMINO-2,6-DIDEOXYGALACTOPYRANOSYL-OXY]-5-(2-OXO-[1,3]DIOXOLAN-4-YL)-3A,5,9,9A-TETRAHYDRO-3H-1-OXA-CYCLOPENTA[A]-S-INDACEN-2-ONE]] | | Authors: | Gao, X, Stassinopoulos, A, Ji, J, Kwon, Y, Bare, S, Goldberg, I.H. | | Deposit date: | 2002-01-26 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Induced formation of a DNA bulge structure by a molecular wedge ligand-postactivated neocarzinostatin chromophore.
Biochemistry, 41, 2002
|
|
