7MJI
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 (focused refinement of RBD and VH ab8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab8 | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
8VLF
 
 | |
6VKJ
 
 | |
7MKM
 
 | |
5IHI
 
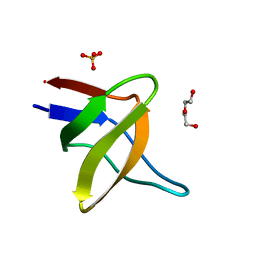 | |
5VWM
 
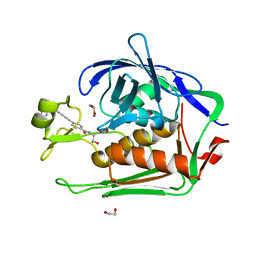 | |
8VC0
 
 | | HIV-1 CA crosslinked pentamer in complex with GS-CA1 | | Descriptor: | Capsid protein p24, N-[(1S)-1-{(3M)-3-{4-chloro-3-[(cyclopropanesulfonyl)amino]-1-(2,2-difluoroethyl)-1H-indazol-7-yl}-6-[3-(methanesulfonyl)-3-methylbut-1-yn-1-yl]pyridin-2-yl}-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-3-(difluoromethyl)-5,5-difluoro-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Piacentini, J, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2023-12-13 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structure of disulfide-stabilized HIV-1 CA pentamer in complex with the ultra-potent capsid inhibitor, GS-CA1
To Be Published
|
|
7MHN
 
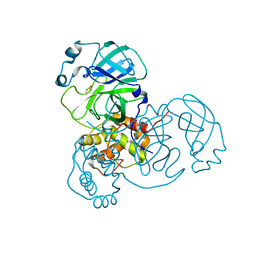 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 277 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1908 Å) | | Cite: | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
8VTC
 
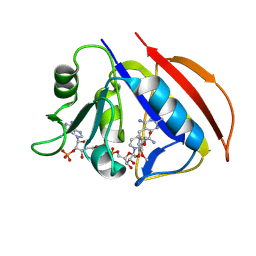 | |
5II1
 
 | | Crystal Structure of the fifth bromodomain of human polybromo (PB1) in complex with 1-methylisochromeno[3,4-c]pyrazol-5(3H)-one | | Descriptor: | 1-methyl[2]benzopyrano[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Myrianthopoulos, V, Mikros, E, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
5VXD
 
 | |
8VDV
 
 | | pcsk9 in complex with inhibitor | | Descriptor: | Inhibitor YBX-PHE-VAL-GLY-THR-THR-PHA-MAA-BIF-EME-NEH, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Xu, M, Chopra, R. | | Deposit date: | 2023-12-18 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | PCSK9 in complex with inhibitor
To Be Published
|
|
7MHO
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 298 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
5VKL
 
 | | SPT6 tSH2-RPB1 1476-1500 pS1493 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Transcription elongation factor SPT6 | | Authors: | Sdano, M.A, Whitby, F.G, Hill, C.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription.
Elife, 6, 2017
|
|
8VLH
 
 | |
7MJL
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (focused refinement of RBD and Fab ab1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, Fab ab1 Light Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
5IJU
 
 | | Structure of an AA10 Lytic Polysaccharide Monooxygenase from Bacillus amyloliquefaciens with Cu(II) bound | | Descriptor: | 1,2-ETHANEDIOL, BaAA10 Lytic Polysaccharide Monooxygenase, CALCIUM ION, ... | | Authors: | Gregory, R.C, Hemsworth, G.R, Turkenburg, J.P, Hart, S.J, Walton, P.H, Davies, G.J. | | Deposit date: | 2016-03-02 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Activity, stability and 3-D structure of the Cu(ii) form of a chitin-active lytic polysaccharide monooxygenase from Bacillus amyloliquefaciens.
Dalton Trans, 45, 2016
|
|
8VI1
 
 | | Crystal structure of c-Met-D1228N in complex with KIN-7615 | | Descriptor: | Hepatocyte growth factor receptor, N-(3,5-difluoro-4-{[6-(2-hydroxyethoxy)-7-methoxyquinolin-4-yl]oxy}phenyl)-4-methoxypyridine-3-carboxamide | | Authors: | Ouyang, X, Kania, R, Cox, J. | | Deposit date: | 2024-01-02 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Discovery of KIN-8741, a Highly Selective Type IIb c-Met Kinase Inhibitor with Broad Mutation Coverage and Quality Drug-Like Properties for the Treatment of Cancer.
J.Med.Chem., 68, 2025
|
|
5IK1
 
 | |
7MHQ
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 310 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9601 Å) | | Cite: | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
8VQJ
 
 | | CryoEM structure of DPOR under turnover | | Descriptor: | COPPER (II) ION, IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein, ... | | Authors: | Kashyap, R, Antony, E. | | Deposit date: | 2024-01-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Cryo-EM captures the coordination of asymmetric electron transfer through a di-copper site in DPOR.
Nat Commun, 16, 2025
|
|
8VZ3
 
 | |
8VYY
 
 | |
6VO6
 
 | | Crystal Structure of Cj1427, an Essential NAD-dependent Dehydrogenase from Campylobacter jejuni, in the Presence of NADH and GDP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Anderson, T.K, Spencer, K.D, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
2Y3P
 
 | | Crystal structure of N-terminal domain of GyrA with the antibiotic simocyclinone D8 | | Descriptor: | DNA GYRASE SUBUNIT A, MAGNESIUM ION, SIMOCYCLINONE D8 | | Authors: | Edwards, M.J, Flatman, R.H, Mitchenall, L.A, Stevenson, C.E.M, Le, T.B.K, Clarke, T.A, McKay, A.R, Fiedler, H.-P, Buttner, M.J, Lawson, D.M, Maxwell, A. | | Deposit date: | 2010-12-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8, Bound to DNA Gyrase.
Science, 326, 2009
|
|
