9B2Q
 
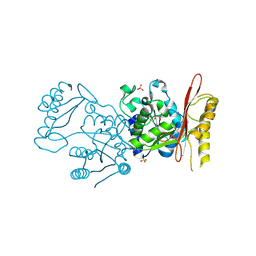 | | Crystal Structure of Pantothenate Synthetase from Candida albicans. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Regan, J, Avad, K.A, Alaidi, O, Seetharaman, J, Palmer, G.E, Hevener, K.E. | | Deposit date: | 2024-03-15 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small molecule inhibitors of fungal pantothenate synthetase can provide a valid and potentially efficacious strategy for antifungal development.
To Be Published
|
|
8X5V
 
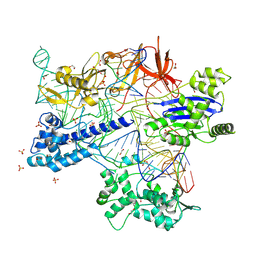 | | BlCas9-sgRNA-target DNA complex | | Descriptor: | 1,2-ETHANEDIOL, BlCas9, CHLORIDE ION, ... | | Authors: | Nakane, T, Nakagawa, R, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2023-11-19 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and engineering of Brevibacillus laterosporus Cas9.
Commun Biol, 7, 2024
|
|
6OK3
 
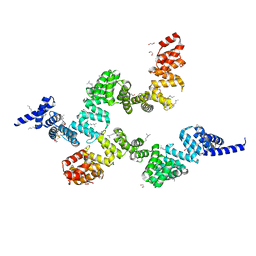 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
8S81
 
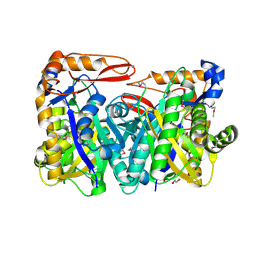 | | VirC mutant C114A-Q334A-R335A-R338A | | Descriptor: | 1,2-ETHANEDIOL, HMG-CoA synthase-like protein | | Authors: | Collin, S, Gruez, A, Weissman, K.J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Plasticity within 3-Hydroxy-3-Methylglutaryl Synthases Catalyzing the First Step of beta-Branching in Polyketide Biosynthesis Underpins a Dynamic Mechanism of Substrate Accommodation.
Jacs Au, 4, 2024
|
|
8V35
 
 | | Crystal structure of HpsN from Cupriavidus pinatubonensis | | Descriptor: | 1,2-ETHANEDIOL, Sulfopropanediol 3-dehydrogenase, ZINC ION | | Authors: | Lee, M. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and kinetic insights into the stereospecific oxidation of R -2,3-dihydroxypropanesulfonate by DHPS-3-dehydrogenase from Cupriavidus pinatubonensis.
Chem Sci, 15, 2024
|
|
4YCD
 
 | |
8V36
 
 | |
5FQI
 
 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
6HXK
 
 | |
7UGM
 
 | |
336D
 
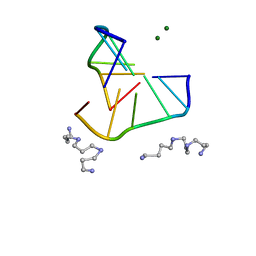 | | INTERACTION BETWEEN LEFT-HANDED Z-DNA AND POLYAMINE-3 THE CRYSTAL STRUCTURE OF THE D(CG)3 AND THERMOSPERMINE COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, N-(3-AMINO-PROPYL)-N-(5-AMINOPROPYL)-1,4-DIAMINOBUTANE | | Authors: | Ohishi, H, Terasoma, N, Nakanishi, I, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Hakoshima, T, Tomita, K.-I. | | Deposit date: | 1997-06-24 | | Release date: | 1998-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between left-handed Z-DNA and polyamine - 3. The crystal structure of the d(CG)3 and thermospermine complex.
FEBS Lett., 398, 1996
|
|
1R3G
 
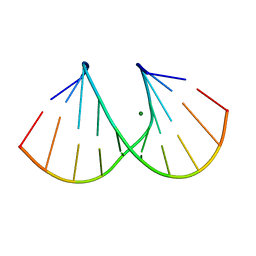 | | 1.16A X-ray structure of the synthetic DNA fragment with the incorporated 2'-O-[(2-Guanidinium)ethyl]-5-methyluridine residues | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(GMU)P*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Prakash, T.P, Puschl, A, Lesnik, E, Tereshko, V, Egli, M, Manoharan, M. | | Deposit date: | 2003-10-01 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 2'-O-[2-(Guanidinium)ethyl]-Modified Oligonucleotides: Stabilizing Effect on Duplex and Triplex Structures.
Org.Lett., 6, 2004
|
|
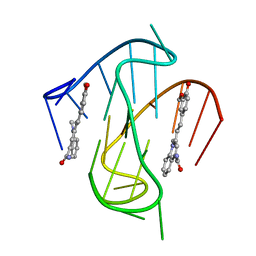
7KBW
 
 | |
4G4M
 
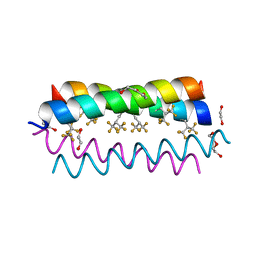 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3(6-13) | | Descriptor: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, alpha4F3(6-13) | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2012-07-16 | | Release date: | 2012-10-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains.
Protein Sci., 21, 2012
|
|
6UZM
 
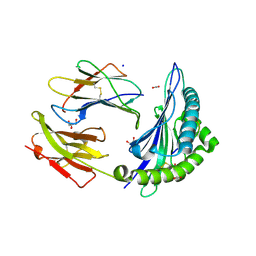 | |
6V6U
 
 | |
4W7R
 
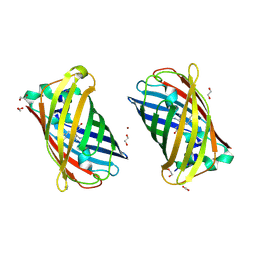 | | Crystal Structure of Full-Length Split GFP Mutant E124H/K126H Copper Mediated Dimer, P 21 Space Group | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, fluorescent protein E124H/K126C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
8BP0
 
 | |
6V64
 
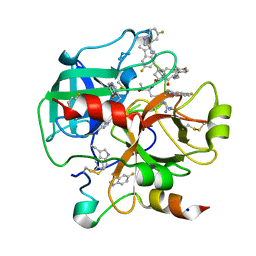 | | Crystal structure of human thrombin bound to ppack with tryptophans replaced by 5-F-tryptophan | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Ruben, E.A, Chen, Z, Di Cera, E. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 19F NMR reveals the conformational properties of free thrombin and its zymogen precursor prethrombin-2.
J.Biol.Chem., 295, 2020
|
|
5NZ3
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, MAGNESIUM ION, RNA (41-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
7Q8A
 
 | | Crystal structure of tandem domain RRM1-2 of FUBP-interacting repressor (FIR) bound to FUSE ssDNA fragment | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*GP*T)-3'), Poly(U)-binding-splicing factor PUF60, ... | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of tandem domain RRM1-2 of FIR bound to FUSE ssDNA fragment
To Be Published
|
|
7A8H
 
 | |
9FCQ
 
 | | CysG(N-16)-H122A mutant in complex with SAH from Kitasatospora cystarginea | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
8SPW
 
 | | PS3 F1 Rotorless, low ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
8YEK
 
 | | Cryo-EM structure of the channelrhodopsin GtCCR2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GtCCR2, RETINAL | | Authors: | Tanaka, T, Iida, W, Sano, F.K, Oda, K, Shihoya, W, Nureki, O. | | Deposit date: | 2024-02-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | The high-light-sensitivity mechanism and optogenetic properties of the bacteriorhodopsin-like channelrhodopsin GtCCR4.
Mol.Cell, 84, 2024
|
|
