6VFP
 
 | | Crystal structure of human protocadherin 1 EC1-EC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin-1, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
3C7O
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with cellotetraose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-08 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
7W3H
 
 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.1_USP14 | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | Deposit date: | 2021-11-25 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
8YHH
 
 | | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Wu, B. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mitotic Kinesin Eg5 from Biortus
To Be Published
|
|
6I8Y
 
 | | Crystal structure of Spindlin1 in complex with the Methyltransferase inhibitor A366 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Shrestha, L, Sorrell, F.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-21 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
8YHS
 
 | | The Crystal Structure of BRDT from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dimethyl-6-[6-(oxan-4-yl)-1-[(1~{S})-1-phenylethyl]imidazo[4,5-c]pyridin-2-yl]pyridazin-3-one, Bromodomain testis-specific protein | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of BRDT from Biortus.
To Be Published
|
|
6V61
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril.
To Be Published
|
|
4TX6
 
 | | AfChiA1 in complex with compound 1 | | Descriptor: | 3-(2-methoxyphenyl)-6-methyl[1,2]oxazolo[5,4-d]pyrimidin-4(5H)-one, Class III chitinase ChiA1, PHOSPHATE ION | | Authors: | van Aalten, D.M.F. | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors
FEBS Lett, 588, 2014
|
|
6HTR
 
 | | Yeast 20S proteasome with human beta2c (S171G) in complex with 13 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, MAGNESIUM ION, PROTEASOME SUBUNIT ALPHA TYPE-1, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6HW3
 
 | | Yeast 20S proteasome in complex with 13 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
4ZG6
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | 4-{(Z)-2-[6-chloro-1-(4-fluorobenzyl)-1H-indol-3-yl]-1-cyanoethenyl}benzoic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
4TYI
 
 | | Structural analysis of the human Fibroblast Growth Factor Receptor 4 | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, Fibroblast growth factor receptor 4 | | Authors: | Lesca, E, Lammens, A, Huber, R, Augustin, M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural analysis of the human fibroblast growth factor receptor 4 kinase.
J.Mol.Biol., 426, 2014
|
|
4TUT
 
 | | Structure of a Prion peptide | | Descriptor: | Prion peptide: GLY-GLY-TYR-MET-LEU-GLY | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-06-24 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
4TZ2
 
 | | Fragment-Based Screening of the Bromodomain of ATAD2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(5-phenyl-4H-1,2,4-triazol-3-yl)aniline, ATPase family AAA domain-containing protein 2, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-09 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
4TYL
 
 | | Fragment-Based Screening of the Bromodomain of ATAD2 | | Descriptor: | 5-amino-1,3,6-trimethyl-1,3-dihydro-2H-benzimidazol-2-one, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-08 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
2KY7
 
 | | NMR Structural Studies on the Covalent DNA Binding of a Pyrrolobenzodiazepine-Naphthalimide Conjugate | | Descriptor: | 2-{2-[4-(3-{[(11aS)-7-methoxy-5-oxo-2,3,5,10,11,11a-hexahydro-1H-pyrrolo[2,1-c][1,4]benzodiazepin-8-yl]oxy}propyl)piperazin-1-yl]ethyl}-1H-benzo[de]isoquinoline-1,3(2H)-dione, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Langel, W, Kamal, A, Weisz, K. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies on the covalent DNA binding of a pyrrolobenzodiazepine-naphthalimide conjugate
Org.Biomol.Chem., 8, 2010
|
|
4UD9
 
 | | Thrombin in complex with 5-chlorothiophene-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-CHLORO-2-THIOPHENECARBOXAMIDE, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2014-12-09 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
4U5B
 
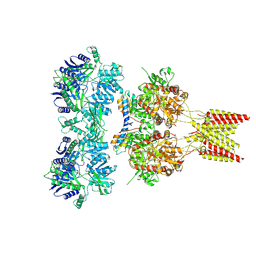 | | Crystal structure of GluA2 A622T, con-ikot-ikot snail toxin, partial agonist KA and postitive modulator (R,R)-2b complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5037 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
8XJI
 
 | | Structure of chimeric RyR complex with flubendiamide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Lin, L, Wang, C, Wang, W, Jiang, H, Yuchi, Z. | | Deposit date: | 2023-12-21 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Cryo-EM structures of ryanodine receptors and diamide insecticides reveal the mechanisms of selectivity and resistance.
Nat Commun, 15, 2024
|
|
4U5E
 
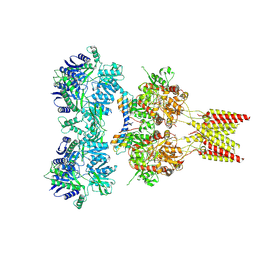 | | Crystal structure of GluA2 T625G, con-ikot-ikot snail toxin, partial agonist KA and postitive modulator (R,R)-2b complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5073 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
4U9A
 
 | |
8X2O
 
 | | RIPK2 in complex with K252 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, ~{N}-[(1~{R})-4-[4-[(6-fluoranyl-1,3-benzothiazol-5-yl)amino]thieno[2,3-d]pyrimidin-6-yl]cyclohex-3-en-1-yl]cyclopropanecarboxamide | | Authors: | Yang, J.H, Yang, J.H. | | Deposit date: | 2023-11-10 | | Release date: | 2024-11-13 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | CMD-OPT model enables the discovery of a potent and selective RIPK2 inhibitor as preclinical candidate for the treatment of acute liver injury.
Acta Pharm Sin B, 15, 2025
|
|
6I44
 
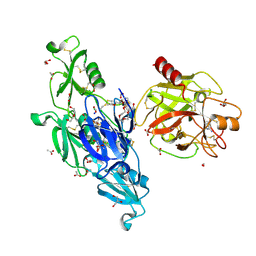 | | Allosteric activation of human prekallikrein by apple domain disc rotation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Li, C, Pathak, M, MaCrae, K, Dreveny, I, Emsley, J. | | Deposit date: | 2018-11-09 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J.Thromb.Haemost., 17, 2019
|
|
6I9A
 
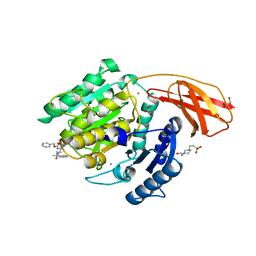 | | Porphyromonas gingivalis gingipain K (Kgp) in complex with inhibitor KYT-36 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gomis-Ruth, F.X, Guevara, T, Rofdriguez-Banqueri, A. | | Deposit date: | 2018-11-22 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural determinants of inhibition of Porphyromonas gingivalis gingipain K by KYT-36, a potent, selective, and bioavailable peptidase inhibitor.
Sci Rep, 9, 2019
|
|
8X0I
 
 | | X-Ray crystal structure of glycoside hydrolase family 6 cellobiohydrolase from Phanerochaete chrysosporium PcCel6A C240S/C393S soaked in cellobioimidazole | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, Glucanase | | Authors: | Yamaguchi, S, Sunagawa, N, Tachioka, M, Igarashi, K. | | Deposit date: | 2023-11-04 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Activity and X-ray crystal structure of candidate base catalyst mutants of glycoside hydrolase family 6 cellobiohydrolase
To Be Published
|
|
