7BNU
 
 | | VDR complex with BXL-62 | | Descriptor: | 1,25-Dihydroxy-16-ene-20-cyclopropyl-vitamin D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2021-01-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Vitamin D Analogs Bearing C-20 Modifications Stabilize the Agonistic Conformation of Non-Responsive Vitamin D Receptor Variants.
Int J Mol Sci, 23, 2022
|
|
7BNS
 
 | | VDR complex with BXL-62 | | Descriptor: | 1,25-Dihydroxy-16-ene-20-cyclopropyl-vitamin D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2021-01-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Vitamin D Analogs Bearing C-20 Modifications Stabilize the Agonistic Conformation of Non-Responsive Vitamin D Receptor Variants.
Int J Mol Sci, 23, 2022
|
|
5OSY
 
 | | Human Decapping Scavenger enzyme (hDcpS) in complex with m7G(5'S)ppSp(5'S)G mRNA 5' cap analog | | Descriptor: | GLYCEROL, [(2~{S},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl-[[[(2~{S},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl-oxidanyl-phosphoryl]oxy-sulfanyl-phosphoryl]oxy-phosphinic acid, m7GpppX diphosphatase | | Authors: | Warminski, M, Nowak, E, Wojtczak, B.A, Fac-Dabrowska, K, Kubacka, D, Nowicka, A, Sikorski, P.J, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 5'-Phosphorothiolate Dinucleotide Cap Analogues: Reagents for Messenger RNA Modification and Potent Small-Molecular Inhibitors of Decapping Enzymes.
J. Am. Chem. Soc., 140, 2018
|
|
5WKK
 
 | | 1.55 A resolution structure of MERS 3CL protease in complex with inhibitor GC813 | | Descriptor: | (1R,2S)-2-[(N-{[2-(3-chlorophenyl)ethoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[2-(3-chlorophenyl)ethoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, MAGNESIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
4NMB
 
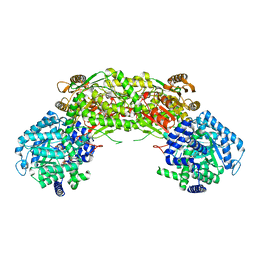 | | Crystal structure of proline utilization A (PutA) from Geobacter sulfurreducens PCA in complex with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Singh, H, Almo, S.C, Tanner, J.J. | | Deposit date: | 2013-11-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5H1E
 
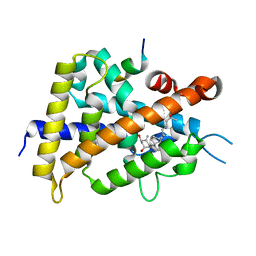 | | Interaction between vitamin D receptor and coactivator peptide SRC2-3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Nuclear receptor coactivator 2 peptide, Vitamin D3 receptor | | Authors: | Egawa, D, Itoh, T, Kato, A, Kataoka, S, Anami, Y, Yamamoto, K. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SRC2-3 binds to vitamin D receptor with high sensitivity and strong affinity
Bioorg. Med. Chem., 25, 2017
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
3HNT
 
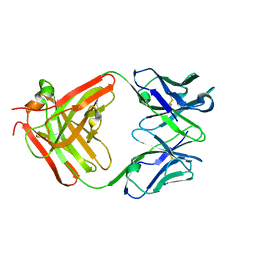 | | CS-35 Fab complex with a linear, terminal oligoarabinofuranosyl tetrasaccharide from lipoarabinomannan | | Descriptor: | CS-35 Fab Heavy Chain, CS-35 Fab Light Chain, beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-5)-alpha-D-arabinofuranose-(1-5)-methyl alpha-D-arabinofuranoside | | Authors: | Murase, T, Zheng, R.B, Joe, M, Bai, Y, Marcus, S.L, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into antibody recognition of mycobacterial polysaccharides.
J.Mol.Biol., 392, 2009
|
|
3QT6
 
 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor DPGP | | Descriptor: | 1-({[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}acetyl)-L-proline, Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
1DRB
 
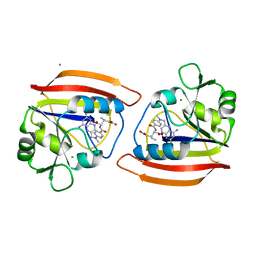 | |
3VPG
 
 | | L-lactate dehydrogenase from Thermus caldophilus GK24 | | Descriptor: | GLYCEROL, L-lactate dehydrogenase | | Authors: | Arai, K, Ohno, T, Miyanaga, A, Fushinobu, S, Taguchi, H. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The core of allosteric motion in Thermus caldophilus L-lactate dehydrogenase.
J.Biol.Chem., 2014
|
|
3W1R
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-045a | | Descriptor: | 2,6-dioxo-5-(2-phenylethyl)-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-20 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-045a
To be Published
|
|
4A29
 
 | | Structure of the engineered retro-aldolase RA95.0 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, D-MALATE, ENGINEERED RETRO-ALDOL ENZYME RA95.0 | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-23 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4NKF
 
 | | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Farnesyl pyrophosphate synthase, ... | | Authors: | Tsoumpra, M.K, Barnett, B.L, Muniz, J.R.C, Walter, R.L, Ebetino, F.H, von Delft, F, Russell, R.G.G, Oppermann, U, Dunford, J.E. | | Deposit date: | 2013-11-12 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates
To be Published
|
|
5TN8
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with (E)-4'-hydroxy-3-((hydroxyiminio)methyl)-[1,1'-biphenyl]-4-olate | | Descriptor: | 3-[(Z)-(hydroxyimino)methyl][1,1'-biphenyl]-4,4'-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Minutolo, F, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
2VWG
 
 | | Haloferax mediterranei glucose dehydrogenase in complex with NADP, Zn and gluconolactone. | | Descriptor: | D-glucono-1,5-lactone, GLUCOSE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Baker, P.J, Britton, K.L, Fisher, M, Esclapez, J, Pire, C, Bonete, M.J, Ferrer, J, Rice, D.W. | | Deposit date: | 2008-06-24 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site dynamics in the zinc-dependent medium chain alcohol dehydrogenase superfamily.
Proc. Natl. Acad. Sci. U.S.A., 106, 2009
|
|
3PRP
 
 | | Structural analysis of a viral OTU domain protease from the Crimean-Congo Hemorrhagic Fever virus in complex with human ubiquitin | | Descriptor: | Polyubiquitin-B (Fragment), RNA-directed RNA polymerase L | | Authors: | Capodagli, G.C, McKercher, M.A, Baker, E.A, Masters, E.M, Brunzelle, J.S, Pegan, S.D. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural analysis of a viral ovarian tumor domain protease from the crimean-congo hemorrhagic Fever virus in complex with covalently bonded ubiquitin.
J.Virol., 85, 2011
|
|
3EUB
 
 | | Crystal Structure of Desulfo-Xanthine Oxidase with Xanthine | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, HYDROXY(DIOXO)MOLYBDENUM, ... | | Authors: | Pauff, J.M, Cao, H, Hille, R. | | Deposit date: | 2008-10-09 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate Orientation and Catalysis at the Molybdenum Site in Xanthine Oxidase: CRYSTAL STRUCTURES IN COMPLEX WITH XANTHINE AND LUMAZINE.
J.Biol.Chem., 284, 2009
|
|
4ZQP
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor MAD1 | | Descriptor: | 5'-O-({1-[(2E)-4-(4-hydroxy-6-methoxy-7-methyl-3-oxo-1,3-dihydro-2-benzofuran-5-yl)-2-methylbut-2-en-1-yl]-1H-1,2,3-triazol-4-yl}methyl)adenosine, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
1OUW
 
 | | Crystal structure of Calystegia sepium agglutinin | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, IMIDAZOLE, ... | | Authors: | Bourne, Y, Roig-Zamboni, V, Barre, A, Peumans, W.J, Astoul, C.H, van Damme, E.J.M, Rouge, P. | | Deposit date: | 2003-03-25 | | Release date: | 2003-11-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The crystal structure of the Calystegia sepium agglutinin reveals a novel quaternary arrangement of lectin subunits with a beta-prism fold
J.Biol.Chem., 279, 2004
|
|
2V2G
 
 | |
6TXR
 
 | | Structural insights into cubane-modified aptamer recognition of a malaria biomarker | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Cheung, Y, Roethlisberger, P, Mechaly, A, Weber, P, Wong, A, Lo, Y, Haouz, A, Savage, P, Hollenstein, M, Tanner, J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1M7I
 
 | | CRYSTAL STRUCTURE OF A MONOCLONAL FAB SPECIFIC FOR SHIGELLA FLEXNERI Y LIPOPOLYSACCHARIDE COMPLEXED WITH A PENTASACCHARIDE | | Descriptor: | alpha-L-rhamnopyranose-(1-2)-alpha-L-rhamnopyranose-(1-3)-alpha-L-rhamnopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-methyl 6-deoxy-alpha-L-rhamnopyranoside, heavy chain of the monoclonal antibody Fab SYA/J6, light chain of the monoclonal antibody Fab SYA/J6 | | Authors: | Vyas, N.K, Vyas, M.N, Chervenak, M.C, Johnson, M.A, Pinto, B.M, Bundle, D.R, Quiocho, F.A. | | Deposit date: | 2002-07-19 | | Release date: | 2003-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Recognition of Oligosaccharide Epitopes by a Monoclonal Fab Specific for
Shigella flexneri Y Lipopolysaccharide: X-ray Structures and Thermodynamics
Biochemistry, 41, 2002
|
|
5H7W
 
 | |
5HCC
 
 | | Ternary complex of human Complement C5 with Ornithodoros moubata OmCI and Dermacentor andersoni RaCI3. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jore, M.M, Johnson, S, Lea, S.M. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for therapeutic inhibition of complement C5.
Nat.Struct.Mol.Biol., 23, 2016
|
|
