6YP2
 
 | |
6YPZ
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
8FIT
 
 | |
6MT1
 
 | |
6PIP
 
 | |
6YOY
 
 | |
6MTA
 
 | |
8T2L
 
 | | Crystal structure of LC3A in complex with the LIR of NSs4 | | Descriptor: | Non-structural protein S,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
6MT5
 
 | |
7RDH
 
 | |
6MVQ
 
 | | HCV NS5B 1b N316 bound to Compound 31 | | Descriptor: | (4-{1-[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl]-1H-1,2,4-triazol-5-yl}-2-fluorophenyl)boronic acid, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
6PBP
 
 | | Pseudopaline Dehydrogenase with (S)-Pseudopaline Soaked 1 hour | | Descriptor: | 1,2-ETHANEDIOL, N-[(1S)-1-carboxy-3-{[(1S)-1-carboxy-2-(1H-imidazol-5-yl)ethyl]amino}propyl]-L-glutamic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
6MYY
 
 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 3 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-02 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
8FR7
 
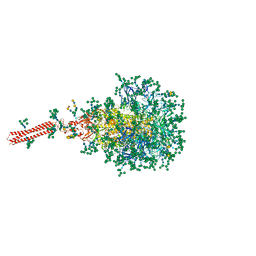 | | A hinge glycan regulates spike bending and impacts coronavirus infectivity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pintilie, G, Wilson, E, Chmielewski, D, Schmid, M.F, Jin, J, Chen, M, Singharoy, A, Chiu, W. | | Deposit date: | 2023-01-06 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural insights into the modulation of coronavirus spike tilting and infectivity by hinge glycans.
Nat Commun, 14, 2023
|
|
7R1O
 
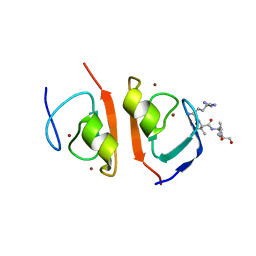 | | p62-ZZ domain of the human sequestosome in complex with dusquetide | | Descriptor: | Dusquetide, Sequestosome-1, ZINC ION | | Authors: | Hakansson, M, Hansson, M, Logan, D.T, Rozek, A, Donini, O. | | Deposit date: | 2022-02-03 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Dusquetide modulates innate immune response through binding to p62.
Structure, 30, 2022
|
|
6N4X
 
 | | Metabotropic Glutamate Receptor 5 Apo Form Ligand Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6PDU
 
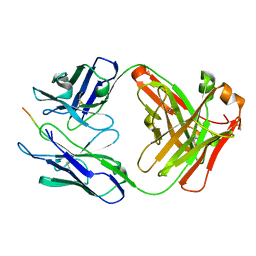 | | Vaccine-elicited NHP FP-targeting antibody 13N024-a.01 in complex with HIV fusion peptide (residue 512-519) | | Descriptor: | HIV-1 fusion peptide residue 512-519, antibody 13N024-a.01, heavy chain, ... | | Authors: | Xu, K, Liu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Modular recognition of antigens provides a mechanism that improves vaccine-elicited antibody-class frequencies
To Be Published
|
|
7RKC
 
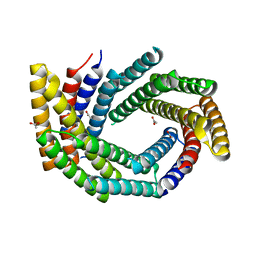 | | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633 | | Descriptor: | ACETATE ION, D_3_633 | | Authors: | Kennedy, M.A, Stoddard, B.L, Hicks, D.R, Bera, A.K. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6PJ8
 
 | |
6N0O
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ523 | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
6PJP
 
 | |
6PRB
 
 | | S. aureus dihydrofolate reductase co-crystallized with cyclopropyl-dimethyoxydihydropthalazine inhibitor and NADP(H) | | Descriptor: | (2E)-1-[(1R)-1-cyclopropyl-6,7-dimethoxyphthalazin-2(1H)-yl]-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}prop-2-en-1-one, (2E)-1-[(1S)-1-cyclopropyl-6,7-dimethoxy-3,4-dihydrophthalazin-2(1H)-yl]-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}prop-2-en-1-one, Dihydrofolate reductase, ... | | Authors: | Bourne, C.R, Thomas, L.M. | | Deposit date: | 2019-07-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor design to target a unique feature in the folate pocket of Staphylococcus aureus dihydrofolate reductase.
Eur.J.Med.Chem., 200, 2020
|
|
7RH2
 
 | | IRF4 Transcription factor mutant -K59R | | Descriptor: | DNA (5'-D(*CP*AP*AP*CP*TP*GP*AP*AP*AP*CP*CP*GP*AP*GP*AP*AP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*TP*TP*CP*TP*CP*GP*GP*TP*TP*TP*CP*AP*GP*TP*TP*G)-3'), ICSAT transcription factor | | Authors: | Williams, S.J, Sundararaj, S, Casarotto, M.G. | | Deposit date: | 2021-07-16 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The molecular basis for the development of adult T-cell leukemia/lymphoma in patients with an IRF4 K59R mutation.
Protein Sci., 31, 2022
|
|
6YI4
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Zak, K.M, Zhou, R.X, Softley, C.A, Bostock, M.J, Sattler, M, Popowicz, G.M. | | Deposit date: | 2020-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion
Not published
|
|
6P9T
 
 | | E.coli LpxA in complex with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc and Compound 8 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
