4S25
 
 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide, S-adenosylhomocysteine, Fe4S4 cluster and Zn (trigonal crystal form) | | Descriptor: | 1,4-BUTANEDIOL, 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
7OFV
 
 | | NMR-guided design of potent and selective EphA4 agonistic ligands | | Descriptor: | ACETATE ION, EphA4 agonist ligand, Ephrin type-A receptor 4 | | Authors: | Ganichkin, O.M, Craig, T.K, Baggio, C, Pellecchia, M. | | Deposit date: | 2021-05-05 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | NMR-Guided Design of Potent and Selective EphA4 Agonistic Ligands.
J.Med.Chem., 64, 2021
|
|
4TRM
 
 | | Structure of the apo form of InhA from Mycobacterium tuberculosis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Chollet, A, Julien, S, Mourey, L, Maveyraud, L. | | Deposit date: | 2014-06-17 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the enoyl-ACP reductase of Mycobacterium tuberculosis (InhA) in the apo-form and in complex with the active metabolite of isoniazid pre-formed by a biomimetic approach.
J.Struct.Biol., 190, 2015
|
|
5YE9
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | N-[4-[(3-cyano-4-naphthalen-2-yloxy-phenyl)sulfamoyl]phenyl]ethanamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
4RKE
 
 | | Drosophila melanogaster Rab2 bound to GMPPNP | | Descriptor: | GH01619p, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Lardong, J.A, Driller, J.H, Depner, H, Weise, C, Petzoldt, A, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | Structures of Drosophila melanogaster Rab2 and Rab3 bound to GMPPNP.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
7NY0
 
 | |
7NXP
 
 | |
7OJ6
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 1 | | Descriptor: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, ACETATE ION, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, ... | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
5Y9T
 
 | | Crystal Structure of EGFR T790M mutant in complex with naquotinib | | Descriptor: | 6-ethyl-3-[[4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-5-[(3R)-1-prop-2-enoylpyrrolidin-3-yl]oxy-pyrazin e-2-carboxamide, Epidermal growth factor receptor | | Authors: | Mimasu, S, Tomimoto, Y, Maiko, I, Yasushi, A, Tatsuya, N. | | Deposit date: | 2017-08-28 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Pharmacological and Structural Characterizations of Naquotinib, a Novel Third-Generation EGFR Tyrosine Kinase Inhibitor, inEGFR-Mutated Non-Small Cell Lung Cancer.
Mol. Cancer Ther., 17, 2018
|
|
5YE7
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | N-[4-[(4-naphthalen-2-yloxyphenyl)sulfamoyl]phenyl]ethanamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
5YGG
 
 | | Crystal structure of Fructokinase Double-Mutant (T261C-H108C) from Vibrio cholerae O395 in fructose, ADP and potassium ion bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Fructokinase, ... | | Authors: | Chatterjee, S, Paul, R, Nath, S, Sen, U. | | Deposit date: | 2017-09-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.671 Å) | | Cite: | Large-scale conformational changes and redistribution of surface negative charge upon sugar binding dictate the fidelity of phosphorylation in Vibrio cholerae fructokinase.
Sci Rep, 8, 2018
|
|
4TRJ
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (INHA) complexed with N-(3-bromophenyl)-1-cyclohexyl-5-oxopyrrolidine-3-carboxamide, refined with new ligand restraints | | Descriptor: | (3S)-N-(3-BROMOPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
5YLL
 
 | | Structure of GH113 beta-1,4-mannanase complex with M6. | | Descriptor: | beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
5YE8
 
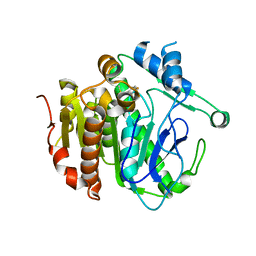 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | N-[3,4-bis(fluoranyl)phenyl]methanesulfonamide, Platelet-activating factor acetylhydrolase | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
5Y3C
 
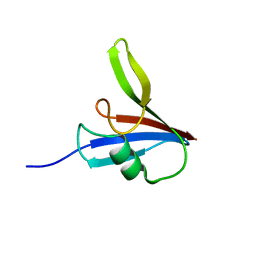 | |
5YQJ
 
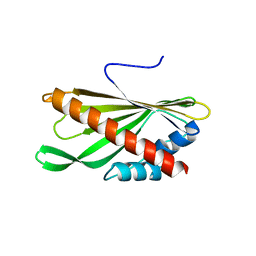 | | Crystal structure of the first StARkin domain of Lam4 | | Descriptor: | Membrane-anchored lipid-binding protein LAM4 | | Authors: | Im, Y.J, Tong, J.S. | | Deposit date: | 2017-11-06 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4TQI
 
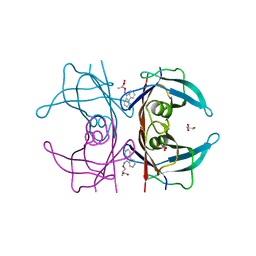 | | Human transthyretin (TTR) complexed with 3-(9H-fluoren-9-ylideneaminooxy)propanoic acid in a dual binding mode | | Descriptor: | (2S)-3-[(9H-fluoren-9-ylideneamino)oxy]-2-methylpropanoic acid, GLYCEROL, Transthyretin | | Authors: | Stura, E.A, Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
4S2X
 
 | | Structure of E. coli RppH bound to RNA and two magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(APC)*GP*U)-3'), RNA pyrophosphohydrolase, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4TT9
 
 | |
5Y3B
 
 | |
4RKG
 
 | | Structure of the MSL2 CXC domain bound with a non-specific (GC)6 DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*C)-3'), E3 ubiquitin-protein ligase msl-2, ZINC ION | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation.
Genes Dev., 28, 2014
|
|
5YEA
 
 | | The crystal structure of Lp-PLA2 in complex with a novel inhibitor | | Descriptor: | 4-[[4-[4-chloranyl-3-(trifluoromethyl)phenoxy]-3-cyano-phenyl]sulfamoyl]benzoic acid, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2017-09-15 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structure-Guided Discovery of Novel, Potent, and Orally Bioavailable Inhibitors of Lipoprotein-Associated Phospholipase A2.
J. Med. Chem., 60, 2017
|
|
5YLI
 
 | | Complex structure of GH113 beta-1,4-mannanase | | Descriptor: | beta-1,4-mannanas, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into the catalytic mechanism of a novel glycoside hydrolase family 113 beta-1,4-mannanase from Amphibacillus xylanus
J. Biol. Chem., 293, 2018
|
|
4RVE
 
 | | THE CRYSTAL STRUCTURE OF ECORV ENDONUCLEASE AND OF ITS COMPLEXES WITH COGNATE AND NON-COGNATE DNA SEGMENTS | | Descriptor: | DNA (5'-D(*GP*GP*GP*AP*TP*AP*TP*CP*CP*C)-3'), PROTEIN (ECO RV (E.C.3.1.21.4)) | | Authors: | Winkler, F.K, Banner, D.W, Oefner, C, Tsernoglou, D, Brown, R.S, Heathman, S.P, Bryan, R.K, Martin, P.D, Petratos, K, Wilso, K.S. | | Deposit date: | 1993-02-18 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments.
EMBO J., 12, 1993
|
|
4RZL
 
 | |
