2C97
 
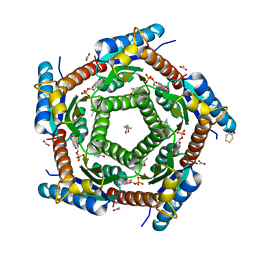 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO 4-(6- chloro-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)butyl phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(6-CHLORO-2,4-DIOXO-1,2,3,4-TETRAHYDROPYRIMIDIN-5-YL) BUTYL PHOSPHATE, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
1ZHS
 
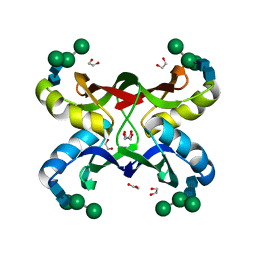 | | Crystal structure of MVL bound to Man3GlcNAc2 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Williams, D.C, Lee, J.Y, Cai, M, Bewley, C.A, Clore, G.M. | | Deposit date: | 2005-04-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the HIV-1 Inhibitory Cyanobacterial Protein MVL Free and Bound to Man3GlcNAc2: STRUCTURAL BASIS FOR SPECIFICITY AND HIGH-AFFINITY BINDING TO THE CORE PENTASACCHARIDE FROM N-LINKED OLIGOMANNOSIDE.
J.Biol.Chem., 280, 2005
|
|
1Z7A
 
 | | Crystal structure of probable Polysaccharide deacetylase from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-24 | | Release date: | 2005-05-10 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of probable Polysaccharide deacetylase from Pseudomonas aeruginosa PAO1
To be Published
|
|
1ZLY
 
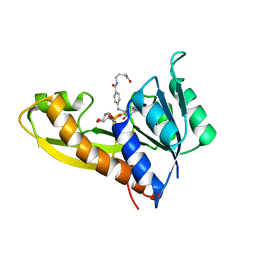 | | The structure of human glycinamide ribonucleotide transformylase in complex with alpha,beta-N-(hydroxyacetyl)-D-ribofuranosylamine and 10-formyl-5,8,dideazafolate | | Descriptor: | 4-[(4-{[(2-AMINO-4-OXO-3,4-DIHYDROQUINAZOLIN-6-YL)METHYL]AMINO}BENZOYL)AMINO]BUTANOIC ACID, 5-O-phosphono-beta-D-ribofuranosylamine, Phosphoribosylglycinamide formyltransferase | | Authors: | Dahms, T.E.S, Sainz, G, Giroux, E.L, Caperelli, C.A, Smith, J.L. | | Deposit date: | 2005-05-09 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The apo and ternary complex structures of a chemotherapeutic target: human glycinamide ribonucleotide transformylase.
Biochemistry, 44, 2005
|
|
2A2O
 
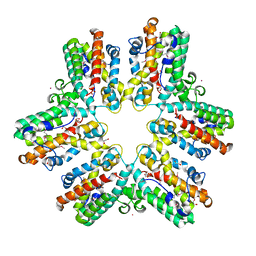 | |
2A8Y
 
 | | Crystal structure of 5'-deoxy-5'methylthioadenosine phosphorylase complexed with 5'-deoxy-5'methylthioadenosine and sulfate | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5'-methylthioadenosine phosphorylase (mtaP), SULFATE ION | | Authors: | Zhang, Y, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2005-07-10 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of 5'-deoxy-5'-methylthioadenosine phosphorylase II from Sulfolobus solfataricus, a thermophilic enzyme stabilized by intramolecular disulfide bonds.
J.Mol.Biol., 357, 2006
|
|
2AUY
 
 | | Pterocarpus angolensis lectin in complex with the trisaccharide GlcNAc(b1-2)Man(a1-3)Man | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-methyl alpha-D-mannopyranoside, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Buts, L, Garcia-Pino, A, Imberty, A, Amiot, N, Boons, G.-J, Beeckmans, S, Versees, W, Wyns, L, Loris, R. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the recognition of complex-type biantennary oligosaccharides by Pterocarpus angolensis lectin.
Febs J., 273, 2006
|
|
1ZLX
 
 | | The apo structure of human glycinamide ribonucleotide transformylase | | Descriptor: | GLYCEROL, Phosphoribosylglycinamide formyltransferase | | Authors: | Dahms, T.E, Sainz, G, Giroux, E.L, Caperelli, C.A, Smith, J.L. | | Deposit date: | 2005-05-09 | | Release date: | 2005-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The apo and ternary complex structures of a chemotherapeutic target: human glycinamide ribonucleotide transformylase.
Biochemistry, 44, 2005
|
|
1ZMC
 
 | | Crystal Structure of Human dihydrolipoamide dehydrogenase complexed to NAD+ | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brautigam, C.A, Chuang, J.L, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal Structure of Human Dihydrolipoamide Dehydrogenase: NAD(+)/NADH Binding and the Structural Basis of Disease-causing Mutations
J.Mol.Biol., 350, 2005
|
|
1ZO8
 
 | | X-ray Structure of the haloalcohol dehalogenase HheC of Agrobacterium radiobacter AD1 in complex with (S)-para-nitrostyrene oxide, with a water molecule in the halide-binding site | | Descriptor: | (S)-PARA-NITROSTYRENE OXIDE, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Tang, L, Villa, A, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-12 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Enantioselectivity of an Epoxide Ring Opening Reaction Catalyzed by Halo Alcohol Dehalogenase HheC.
J.Am.Chem.Soc., 127, 2005
|
|
1ZF3
 
 | | ATC Four-stranded DNA Holliday Junction | | Descriptor: | 5'-D(*CP*CP*GP*AP*TP*AP*TP*CP*GP*G)-3', CALCIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZFH
 
 | | TTA Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*TP*AP*AP*TP*TP*AP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AAM
 
 | |
2CE8
 
 | |
2C9B
 
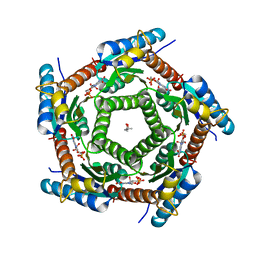 | | Lumazine Synthase from Mycobacterium tuberculosus Bound to 3-(1,3,7- TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(1,3,7-TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL), 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
2CJF
 
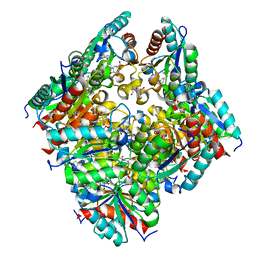 | | TYPE II DEHYDROQUINASE INHIBITOR COMPLEX | | Descriptor: | (1S,4S,5S)-1,4,5-TRIHYDROXY-3-[3-(PHENYLTHIO)PHENYL]CYCLOHEX-2-ENE-1-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Payne, R.J, Riboldi-Tunnicliffe, A, Abell, A.D, Lapthorn, A.J, Abell, C. | | Deposit date: | 2006-03-31 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis, and structural studies on potent biaryl inhibitors of type II dehydroquinases.
Chemmedchem, 2, 2007
|
|
2BC1
 
 | |
2BC0
 
 | |
2BCP
 
 | |
2C91
 
 | | mouse succinic semialdehyde reductase, AKR7A5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AFLATOXIN B1 ALDEHYDE REDUCTASE MEMBER 2, GLYCEROL, ... | | Authors: | Zhu, X, Ellis, E.M, Lapthorn, A.J. | | Deposit date: | 2005-12-08 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Mouse Succinic Semialdehyde Reductase Akr7A5: Structural Basis for Substrate Specificity.
Biochemistry, 45, 2006
|
|
2CA9
 
 | | apo-NIKR from helicobacter pylori in closed trans-conformation | | Descriptor: | FORMIC ACID, GLYCEROL, PUTATIVE NICKEL-RESPONSIVE REGULATOR | | Authors: | Dian, C, Schauer, K, Kapp, U, McSweeney, S.M, Labigne, A, Terradot, L. | | Deposit date: | 2005-12-20 | | Release date: | 2006-07-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of the Nickel Response in Helicobacter Pylori: Crystal Structures of Hpnikr in Apo and Nickel-Bound States.
J.Mol.Biol., 361, 2006
|
|
2CAD
 
 | | NikR from Helicobacter pylori in closed trans-conformation and nickel bound to 2F, 2X and 2I sites. | | Descriptor: | CITRIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Dian, C, Schauer, K, Kapp, U, McSweeney, S.M, Labigne, A, Terradot, L. | | Deposit date: | 2005-12-20 | | Release date: | 2006-07-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Nickel Response in Helicobacter Pylori: Crystal Structures of Hpnikr in Apo and Nickel-Bound States.
J.Mol.Biol., 361, 2006
|
|
2CJT
 
 | | Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Heterodimer Switch: C2-domains as Versatile Protein-Protein Interaction Modules | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, UNC-13 HOMOLOG A | | Authors: | Lu, J, Machius, M, Dulubova, I, Dai, H, Sudhof, T.C, Tomchick, D.R, Rizo, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for a Munc13-1 Dimeric to Munc13-1/Rim Heterodimer Switch
Plos Biol., 4, 2006
|
|
2CMP
 
 | | crystal structure of the DNA binding domain of G1P SMALL TERMINASE SUBUNIT from bacteriophage SF6 | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Benini, S, Chechik, M, Ortiz-Lombardia, M, Polier, S, Shevtsov, M.B, DeLuchi, D, Alonso, J.C, Antson, A.A. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The 1.58 A Resolution Structure of the DNA-Binding Domain of Bacteriophage Sf6 Small Terminase Provides New Hints on DNA Binding
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2D8P
 
 | | Structure of HYPER-VIL-thaumatin | | Descriptor: | IODIDE ION, Thaumatin I | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-08 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
