5FGB
 
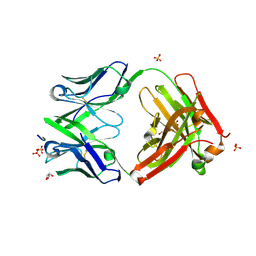 | | Three dimensional structure of broadly neutralizing human anti - Hepatitis C virus (HCV) glycoprotein E2 Fab fragment HC33.4 | | Descriptor: | Anti-HCV E2 Fab HC84-1 heavy chain, Anti-HCV E2 Fab HC84-1 light chain, GLYCEROL, ... | | Authors: | Girard-Blanc, C, Rey, F.A, Krey, T. | | Deposit date: | 2015-12-20 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Antibody Response to Hypervariable Region 1 Interferes with Broadly Neutralizing Antibodies to Hepatitis C Virus.
J.Virol., 90, 2016
|
|
5HFN
 
 | |
4QKS
 
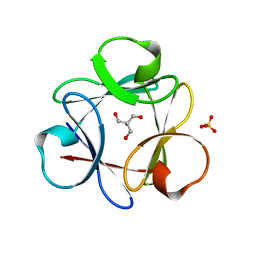 | | Crystal Structure of 6xTrp/PV2: de novo designed beta-trefoil architecture with symmetric primary structure (L22W/L44W/L64W/L85W/L108W/L132W his Primitive Version 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DE NOVO PROTEIN 6XTRP/PV2, SULFATE ION | | Authors: | Longo, L.M, Tenorio, C.A, Blaber, M. | | Deposit date: | 2014-06-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A single aromatic core mutation converts a designed "primitive" protein from halophile to mesophile folding.
Protein Sci., 24, 2015
|
|
5HAQ
 
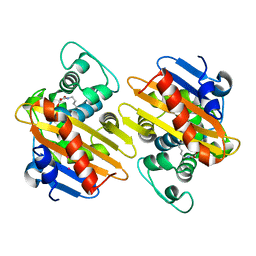 | | OXa-48 beta-lactamase mutant - S70G | | Descriptor: | Beta-lactamase, CADMIUM ION, FORMIC ACID | | Authors: | Stojanoski, V, Adamski, C.J, Hu, L, Mehta, S.C, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2015-12-30 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Removal of the Side Chain at the Active-Site Serine by a Glycine Substitution Increases the Stability of a Wide Range of Serine beta-Lactamases by Relieving Steric Strain.
Biochemistry, 55, 2016
|
|
5HAP
 
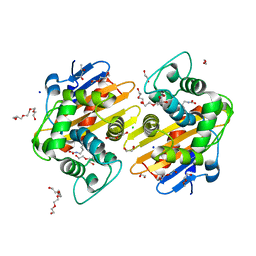 | | OXA-48 beta-lactamase - S70A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Stojanoski, V, Adamski, C.J, Hu, L, Mehta, S.C, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2015-12-30 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Removal of the Side Chain at the Active-Site Serine by a Glycine Substitution Increases the Stability of a Wide Range of Serine beta-Lactamases by Relieving Steric Strain.
Biochemistry, 55, 2016
|
|
5HDB
 
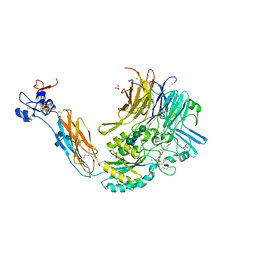 | | Integrin alphaIIbbeta3 in complex with Ro-435054 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lin, F.Y. | | Deposit date: | 2016-01-05 | | Release date: | 2016-02-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7012 Å) | | Cite: | beta-Subunit Binding Is Sufficient for Ligands to Open the Integrin alpha IIb beta 3 Headpiece.
J.Biol.Chem., 291, 2016
|
|
3RMD
 
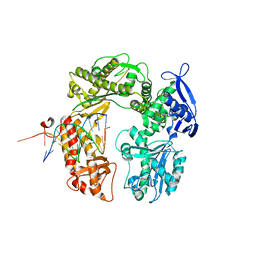 | | Crystal Structure of a replicative DNA polymerase bound to DNA containing Thymine Glycol | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*GP*TP*(CTG)P*G*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*CP*AP*A)-3'), ... | | Authors: | Aller, P, Duclos, S, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A crystallographic study of the role of sequence context in thymine glycol bypass by a replicative DNA polymerase serendipitously sheds light on the exonuclease complex.
J.Mol.Biol., 412, 2011
|
|
4TKS
 
 | |
4TKR
 
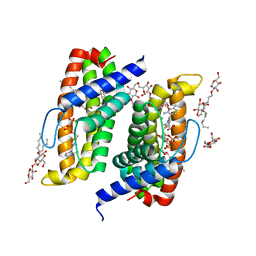 | | Native-SAD phasing for ThiT from Listeria monocytogenes serovar. | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, THIAMINE DIPHOSPHATE, Thiamine transporter thia | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.0023 Å) | | Cite: | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TKQ
 
 | | Native-SAD phasing for YetJ from Bacillus Subtilis | | Descriptor: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein YetJ | | Authors: | Liu, Q, Chang, Y, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8025 Å) | | Cite: | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TXY
 
 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase, a prokaryotic cGAS homolog | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.0001 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
3PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (QB-DEPLETED) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB.
Structure, 5, 1997
|
|
2LJD
 
 | |
2LJF
 
 | |
2LOY
 
 | | Refined Miminal Constraint Solution NMR Structure of Translationally-controlled tumor protein (TCTP) from Caenorhabditis elegans, Northeast Structural Genomics Consortium Target WR73 | | Descriptor: | Translationally-controlled tumor protein homolog | | Authors: | Aramini, J.M, Rossi, P, Cort, J.R, Lee, H, Janjua, H, Maglaqui, M, Cooper, B, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6QZ1
 
 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | BENZOIC ACID, CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2LQX
 
 | | NMR spatial structure of the trypsin inhibitor BWI-2c from the buckwheat seeds | | Descriptor: | Trypsin inhibitor BWI-2c | | Authors: | Mineev, K.S, Vassilevski, A.A, Oparin, P.B, Grishin, E.V, Egorov, T.A. | | Deposit date: | 2012-03-19 | | Release date: | 2012-05-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Buckwheat trypsin inhibitor with helical hairpin structure belongs to a new family of plant defence peptides.
Biochem.J., 446, 2012
|
|
2LMZ
 
 | |
2M1R
 
 | | PHD domain of ING4 N214D mutant | | Descriptor: | Inhibitor of growth protein 4, ZINC ION | | Authors: | Blanco, F.J. | | Deposit date: | 2012-12-05 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Functional impact of cancer-associated mutations in the tumor suppressor protein ING4.
Carcinogenesis, 31, 2010
|
|
7SD5
 
 | | Crystallographic structure of neutralizing antibody 10-40 in complex with SARS-CoV-2 spike receptor binding domain | | Descriptor: | 10-40 Heavy chain, 10-40 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Reddem, E.R, Casner, R.G, Shapiro, L. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses.
Sci Transl Med, 14, 2022
|
|
7SI2
 
 | |
2M97
 
 | |
2LZO
 
 | | Spatial structure of Pi-AnmTX Ugr 9a-1 | | Descriptor: | UGTX | | Authors: | Mineev, K, Arseniev, A. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Sea Anemone Peptide with Uncommon beta-Hairpin Structure Inhibits Acid-sensing Ion Channel 3 (ASIC3) and Reveals Analgesic Activity.
J.Biol.Chem., 288, 2013
|
|
6RR5
 
 | | Structure of 50% reduced KpDyP | | Descriptor: | GLYCEROL, Iron-dependent peroxidase, MAGNESIUM ION, ... | | Authors: | Pfanzagl, V, Beale, J, Hofbauer, S. | | Deposit date: | 2019-05-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray-induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner.
J.Biol.Chem., 295, 2020
|
|
4L7I
 
 | | Crystal structure of S-Adenosylmethionine synthase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
