6ZY9
 
 | | Cryo-EM structure of MlaFEDB in complex with AMP-PNP | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, MAGNESIUM ION, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7P73
 
 | | The PDZ domain of SYNJ2BP complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | CALCIUM ION, GLYCEROL, Protein Tax-1, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
7U5O
 
 | | CRYSTAL STRUCTURE OF THE BONE MORPHOGENETIC PROTEIN RECEPTOR TYPE 2 LIGAND BINDING DOMAIN IN COMPLEX WITH ACTIVIN-B | | Descriptor: | Bone morphogenetic protein receptor type-2, Inhibin beta B chain | | Authors: | Chu, K.Y, Malik, A, Thamilselvan, V, Martinez-Hackert, E. | | Deposit date: | 2022-03-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Type II BMP and activin receptors BMPR2 and ACVR2A share a conserved mode of growth factor recognition.
J.Biol.Chem., 298, 2022
|
|
3HU2
 
 | | Structure of p97 N-D1 R86A mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.-K. | | Deposit date: | 2009-06-12 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
7EJ2
 
 | | human voltage-gated potassium channel KV1.3 H451N mutant | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 3, Voltage-gated potassium channel subunit beta-2 | | Authors: | Liu, S, Zhao, Y, Tian, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of wild-type and H451N mutant human lymphocyte potassium channel K V 1.3.
Cell Discov, 7, 2021
|
|
7R86
 
 | | Structure of mouse BAI1 (ADGRB1) in complex with mouse Nogo receptor (RTN4R) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINOBENZOIC ACID, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-06-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development.
Cell, 184, 2021
|
|
7ECF
 
 | | Crystal Structure of d(G4C2)2-Ba in C2221 space group | | Descriptor: | BARIUM ION, DNA (5'-D(*GP*GP*GP*GP*CP*CP*GP*GP*GP*GP*CP*C)-3') | | Authors: | Geng, Y, Liu, C, Cai, Q, Luo, Z, Zhu, G. | | Deposit date: | 2021-03-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of parallel G-quadruplex formed by the two-repeat ALS- and FTD-related GGGGCC sequence.
Nucleic Acids Res., 49, 2021
|
|
2JGI
 
 | |
7LY7
 
 | | Crystal structure of the elongation module of the bacillamide NRPS, BmdB, in complex with the oxidase BmdC | | Descriptor: | 5'-{[(2R,3S)-3-amino-2-({2-[(N-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-beta-alanyl)amino]ethyl}sulfanyl)-4-sulfanylbutane-1-sulfonyl]amino}-5'-deoxyadenosine, BmdB, Bacillamide NRPS, ... | | Authors: | Fortinez, C.M, Sharon, I, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
7EJ1
 
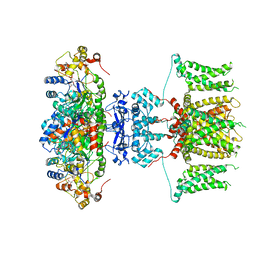 | | human voltage-gated potassium channel KV1.3 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 3, Voltage-gated potassium channel subunit beta-2 | | Authors: | Liu, S, Zhao, Y, Tian, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of wild-type and H451N mutant human lymphocyte potassium channel K V 1.3.
Cell Discov, 7, 2021
|
|
3HW9
 
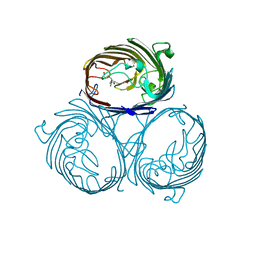 | | Cation selective pathway of OmpF porin revealed by anomalous x-ray diffraction | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Outer membrane protein F, ... | | Authors: | Balasundaresan, D, Raychaudhury, S, Blachowicz, L, Roux, B. | | Deposit date: | 2009-06-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Cation-selective pathway of OmpF porin revealed by anomalous X-ray diffraction.
J.Mol.Biol., 396, 2010
|
|
2ZHX
 
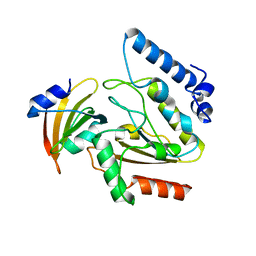 | | Crystal structure of Uracil-DNA Glycosylase from Mycobacterium tuberculosis in complex with a proteinaceous inhibitor | | Descriptor: | Uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | Authors: | Kaushal, P.S, Talawar, R.K, Krishna, P.D.V, Varshney, U, Vijayan, M. | | Deposit date: | 2008-02-11 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Unique features of the structure and interactions of mycobacterial uracil-DNA glycosylase: structure of a complex of the Mycobacterium tuberculosis enzyme in comparison with those from other sources
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7R85
 
 | | Structure of mouse Bai1 (ADGRB1) TSR3 domain | | Descriptor: | Vasculostatin-120, alpha-D-mannopyranose, beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-06-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | RTN4/NoGo-receptor binding to BAI adhesion-GPCRs regulates neuronal development.
Cell, 184, 2021
|
|
7TL3
 
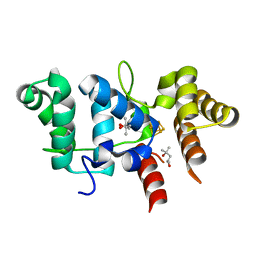 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF431 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, IRON/SULFUR CLUSTER | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.066 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
8R1T
 
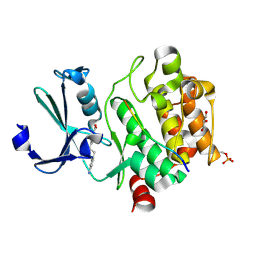 | | Pim1 in complex with 4-(4-aminophenethyl)benzoic acid and Pimtide | | Descriptor: | 4-(4-aminophenethyl)benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | What doesn't fit is made to fit: Pim-1 kinase adapts to the configuration of stilbene-based inhibitors.
Arch Pharm, 357, 2024
|
|
7LBC
 
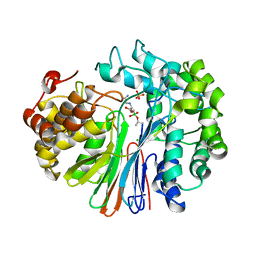 | | Structure of human GGT1 in complex with Lnt2-65 compound | | Descriptor: | (2R)-2-[(2-aminoethyl)amino]-4-boronobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2021-01-07 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Design and evaluation of novel analogs of 2-amino-4-boronobutanoic acid (ABBA) as inhibitors of human gamma-glutamyl transpeptidase.
Bioorg.Med.Chem., 73, 2022
|
|
2OC4
 
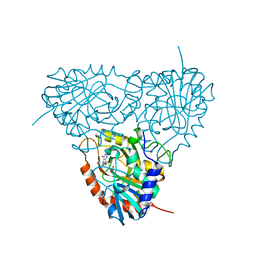 | | Crystal structure of human purine nucleoside phosphorylase mutant H257D with Imm-H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2006-12-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Neighboring Group Participation in the Transition State of Human Purine Nucleoside Phosphorylase
Biochemistry, 46, 2007
|
|
7EF9
 
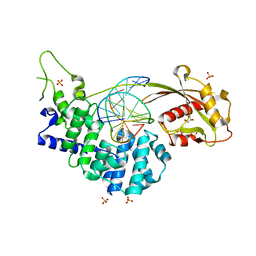 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form II) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*AP*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
5M0Z
 
 | |
7TH1
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B3 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-7-benzyl-4-[(3-methylphenyl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7E9G
 
 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu2 | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, DN13, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
2JIX
 
 | | Crystal structure of ABT-007 FAB fragment with the soluble domain of EPO receptor | | Descriptor: | ABT-007 FAB FRAGMENT, ERYTHROPOIETIN RECEPTOR | | Authors: | Liu, Z, Stoll, V.S, DeVries, P, Jakob, C.G, Xie, N, Simmer, R.L, Lacy, S.E, Egan, D.A, Harlan, J.E, Lesniewski, R.R, Reilly, E.B. | | Deposit date: | 2007-07-02 | | Release date: | 2007-07-10 | | Last modified: | 2020-03-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Potent Erythropoietin-Mimicking Human Antibody Interacts Through a Novel Binding Site.
Blood, 110, 2007
|
|
2JVL
 
 | | NMR structure of the C-terminal domain of MBF1 of Trichoderma reesei | | Descriptor: | TrMBF1 | | Authors: | Kopke Salinas, R, Tomaselli, S, Camilo, C.M, Valencia, E.Y, Farah, C.S, El-Dorry, H, Chambergo, F.S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of multiprotein bridging factor 1 (MBF1) of Trichoderma reesei.
Proteins, 75, 2009
|
|
7EF8
 
 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form I) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
5M2G
 
 | | PCE reductive dehalogenase from S. multivorans in complex with 2,4,6-tribromophenol | | Descriptor: | 2,4,6-TRIBROMOPHENOL, BENZAMIDINE, GLYCEROL, ... | | Authors: | Kunze, C, Bommer, M, Hagen, W.R, Uksa, M, Dobbek, H, Schubert, T, Diekert, G. | | Deposit date: | 2016-10-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer.
Nat Commun, 8, 2017
|
|
