4LGA
 
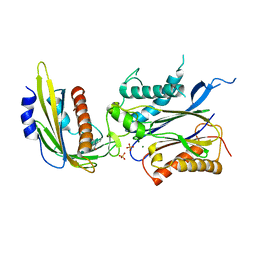 | | ABA-mimicking ligand N-(2-OXO-1-PROPYL-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-PHENYLMETHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)-1-phenylmethanesulfonamide, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
1NJS
 
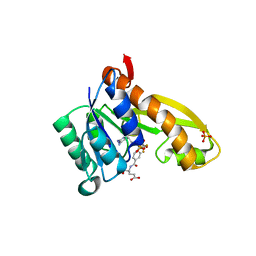 | | human GAR Tfase in complex with hydrolyzed form of 10-trifluoroacetyl-5,10-dideaza-acyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Marsilje, T.H, Li, C, Hedrick, M.P, Gooljarsingh, L.T, Tavassoli, A, Benkovic, S.J, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Rational Design, Synthesis, Evaluation, and Crystal Structure of a Potent Inhibitor of Human GAR Tfase: 10-(Trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic Acid
Biochemistry, 42, 2003
|
|
5SA9
 
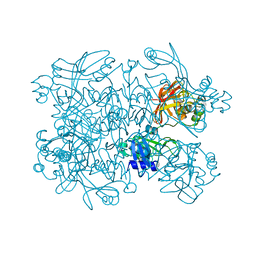 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2697514548 | | Descriptor: | 1-methylindazole-3-carboxamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
2HXQ
 
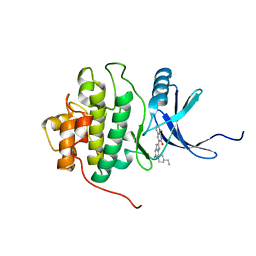 | | crystal structure of Chek1 in complex with inhibitor 2 | | Descriptor: | 3-(5-{[4-(AMINOMETHYL)PIPERIDIN-1-YL]METHYL}-1H-INDOL-2-YL)QUINOLIN-2(1H)-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of 6-substituted indolylquinolinones as potent Chek1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3FVS
 
 | | Human Kynurenine Aminotransferase I in complex with Glycerol | | Descriptor: | GLYCEROL, Kynurenine--oxoglutarate transaminase 1, SODIUM ION | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
3BOV
 
 | | Crystal structure of the receptor binding domain of mouse PD-L2 | | Descriptor: | FORMIC ACID, Programmed cell death 1 ligand 2, SODIUM ION | | Authors: | Lazar-Molnar, E, Ramagopal, U, Cao, E, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-17 | | Release date: | 2008-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1BU1
 
 | | SRC FAMILY KINASE HCK SH3 DOMAIN | | Descriptor: | PROTEIN (HEMOPOIETIC CELL KINASE) | | Authors: | Arold, S, Franken, P, Dumas, C. | | Deposit date: | 1998-09-09 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef.
Biochemistry, 37, 1998
|
|
4B2D
 
 | | human PKM2 with L-serine and FBP bound. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PYRUVATE KINASE ISOZYMES M1/M2, ... | | Authors: | Chaneton, B, Hillmann, P, Zheng, L, Martin, A.C.L, Maddocks, O.D.K, Chokkathukalam, A, Coyle, J.E, Jankevics, A, Holding, F.P, Vousden, K.H, Frezza, C, O'Reilly, M, Gottlieb, E. | | Deposit date: | 2012-07-13 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serine is a natural ligand and allosteric activator of pyruvate kinase M2.
Nature, 491, 2012
|
|
2Y4S
 
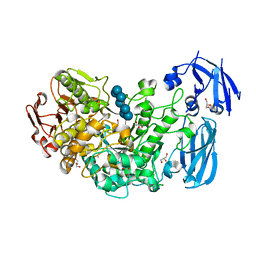 | | BARLEY LIMIT DEXTRINASE IN COMPLEX WITH BETA-CYCLODEXTRIN | | Descriptor: | CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Vester-Christensen, M.B, Hachem, M.A, Svensson, B, Henriksen, A. | | Deposit date: | 2011-01-10 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of an Essential Enzyme in Seed Starch Degradation: Barley Limit Dextrinase in Complex with Cyclodextrins.
J.Mol.Biol., 403, 2010
|
|
3MVM
 
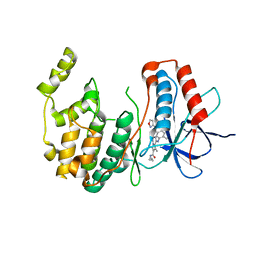 | | P38 Alpha Map Kinase complexed with pyrrolotriazine inhibitor 7V | | Descriptor: | 4-{[5-(isoxazol-3-ylcarbamoyl)-2-methylphenyl]amino}-5-methyl-N-propylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-(5-(Cyclopropylcarbamoyl)-2-Methylphenylamino)-5-Methyl-Npropylpyrrolo[1,2-F][1,2,4] Triazine-6-Carboxamide (Bms-582949), a Clinical P38? Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
J.Med.Chem., 53, 2010
|
|
2B7Z
 
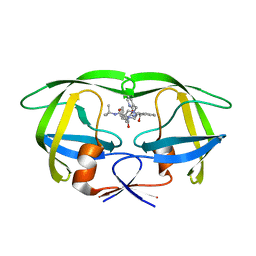 | | Structure of HIV-1 protease mutant bound to indinavir | | Descriptor: | HIV-1 protease, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE | | Authors: | Clemente, J.C, Stow, L.R, Janka, L.K, Jeung, J.A, Desai, K.A, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2005-10-05 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vivo, kinetic, and structural analysis of the development of ritonavir resistance
To be Published
|
|
3WEH
 
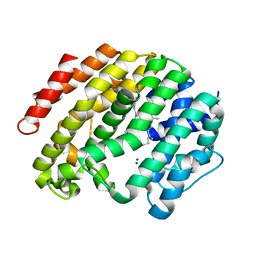 | | Crystal structure of the human squalene synthase in complex with presqualene pyrophosphate | | Descriptor: | MAGNESIUM ION, Squalene synthase, {(1R,2R,3R)-2-[(3E)-4,8-dimethylnona-3,7-dien-1-yl]-2-methyl-3-[(1E,5E)-2,6,10-trimethylundeca-1,5,9-trien-1-yl]cyclopropyl}methyl trihydrogen diphosphate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-07-07 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insights into the catalytic mechanism of human squalene synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3R7O
 
 | | Structure of dually phosphorylated c-MET receptor kinase in complex with an MK-2461 analog | | Descriptor: | Hepatocyte growth factor receptor, N-[(2R)-1,4-dioxan-2-ylmethyl]-N-methyl-N'-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}sulfuric diamide | | Authors: | Soisson, S.M, Rickert, K, Patel, S.B, Munshi, S, Lumb, K.J. | | Deposit date: | 2011-03-22 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for selective small molecule kinase inhibition of activated c-Met.
J.Biol.Chem., 286, 2011
|
|
4HB2
 
 | | Crystal structure of CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2B1A
 
 | | Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with UG1033 peptide | | Descriptor: | Fab 2219, heavy chain, light chain, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity.
J.Virol., 80, 2006
|
|
2B2F
 
 | |
4HCR
 
 | |
5Z57
 
 | | Cryo-EM structure of the human activated spliceosome (late Bact) at 6.5 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ALANINE, BUD13 homolog, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
1W1G
 
 | | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain bound to DiC4-phosphatidylinositol (3,4,5)-trisphosphate | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1 | | Authors: | Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-06-21 | | Release date: | 2004-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Insights Into the Regulation of Pdk1 by Phosphoinositides and Inositol Phosphates
Embo J., 23, 2004
|
|
4HAU
 
 | |
5RGO
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102248 (Mpro-x0736) | | Descriptor: | 1-[4-(furan-2-carbonyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
2Y5E
 
 | | BARLEY LIMIT DEXTRINASE IN COMPLEX WITH ALPHA-CYCLODEXTRIN | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Vester-Christensen, M.B, Hachem, M.A, Svensson, B, Henriksen, A. | | Deposit date: | 2011-01-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal Structure of an Essential Enzyme in Seed Starch Degradation: Barley Limit Dextrinase in Complex with Cyclodextrins.
J.Mol.Biol., 403, 2010
|
|
5RFU
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102121 | | Descriptor: | 1-{4-[(5-chlorothiophen-2-yl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5ZH1
 
 | |
7KU7
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. Cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
