5TCG
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate-bound form | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, FORMIC ACID, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
5TEN
 
 | | Structure of 4-Hydroxy-tetrahydrodipicolinate Reductase from Vibrio vulnificus with 2,5 Furan Dicarboxylic and NADH with Intact Polyhistidine Tag | | Descriptor: | 2,5 Furan Dicarboxylic Acid, 4-hydroxy-tetrahydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mank, N, Pote, S, Arnette, K, Klapper, V, Chruszcz, M. | | Deposit date: | 2016-09-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6RM3
 
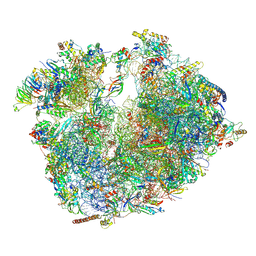 | | Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | Authors: | Barandun, J, Hunziker, M, Vossbrinck, C.R, Klinge, S. | | Deposit date: | 2019-05-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome.
Nat Microbiol, 4, 2019
|
|
5TIG
 
 | |
6OC7
 
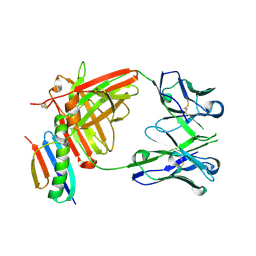 | | HMP42 Fab in complex with Protein G | | Descriptor: | Heavy chain of HMP42 Fab, Immunoglobulin G-binding protein G, Light chain for HMP42 Fab | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2019-03-22 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
6RPB
 
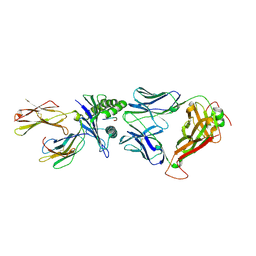 | | Crystal structure of the T-cell receptor NYE_S1 bound to HLA A2*01-SLLMWITQV | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Coles, C.H, Mulvaney, R, Malla, S, Lloyd, A, Smith, K, Chester, F, Knox, A, Stacey, A.R, Dukes, J, Baston, E, Griffin, S, Vuidepot, A, Jakobsen, B.K, Harper, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | TCRs with Distinct Specificity Profiles Use Different Binding Modes to Engage an Identical Peptide-HLA Complex.
J Immunol., 204, 2020
|
|
4XGS
 
 | |
6OFR
 
 | |
6ROL
 
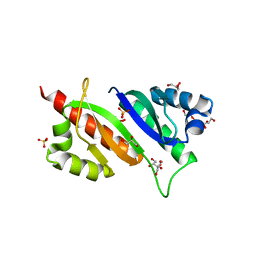 | | Structure of IMP2 KH34 domains | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Insulin-like growth factor 2 mRNA-binding protein 2, ... | | Authors: | Bhaskar, V, Biswas, J, Singer, R.H, Chao, J.A. | | Deposit date: | 2019-05-13 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for RNA selectivity by the IMP family of RNA-binding proteins.
Nat Commun, 10, 2019
|
|
6OI8
 
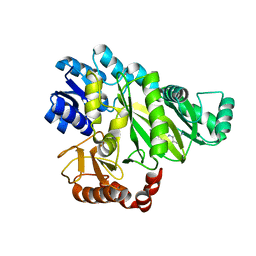 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with 7-((1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl)-6-(2-chloro-6-(pyridin-3-yl)phenyl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 7-[(1R,5S,6s)-6-amino-3-azabicyclo[3.1.0]hexan-3-yl]-6-[2-chloro-6-(pyridin-3-yl)phenyl]pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
6RVY
 
 | |
5THT
 
 | | Crystal Structure of G303A HDAC8 in complex with M344 | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
6RUV
 
 | |
6ONG
 
 | | Dehaloperoxidate B in complex with substrate 4-F-cresol | | Descriptor: | 1,2-ETHANEDIOL, 4-fluoro-2-methylphenol, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T, McGuire, A. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The multifunctional globin dehaloperoxidase strikes again: Simultaneous peroxidase and peroxygenase mechanisms in the oxidation of EPA pollutants.
Arch.Biochem.Biophys., 673, 2019
|
|
6RXT
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state A | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6IP5
 
 | | Cryo-EM structure of the CMV-stalled human 80S ribosome (Structure ii) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6IM3
 
 | | Crystal structure of a highly thermostable carbonic anhydrase from Persephonella marina EX-H1 | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CALCIUM ION, Carbonic anhydrase (Carbonate dehydratase), ... | | Authors: | Jin, M.S, Kim, S, Sung, J, Yeon, J, Choi, S.H. | | Deposit date: | 2018-10-22 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Highly Thermostable alpha-Carbonic Anhydrase from Persephonella marina EX-H1.
Mol.Cells, 42, 2019
|
|
4XDS
 
 | | Deoxyguanosinetriphosphate Triphosphohydrolase from Escherichia coli with Nickel | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, NICKEL (II) ION, SULFATE ION | | Authors: | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | Deposit date: | 2014-12-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
5T1O
 
 | | Solution-state NMR and SAXS structural ensemble of NPr (1-85) in complex with EIN-Ntr (170-424) | | Descriptor: | Phosphocarrier protein NPr, Phosphoenolpyruvate-protein phosphotransferase PtsP | | Authors: | Strickland, M, Stanley, A.M, Wang, G, Schwieters, C.D, Buchanan, S, Peterkofsky, A, Tjandra, N. | | Deposit date: | 2016-08-19 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure of the NPr:EIN(Ntr) Complex: Mechanism for Specificity in Paralogous Phosphotransferase Systems.
Structure, 24, 2016
|
|
6RXY
 
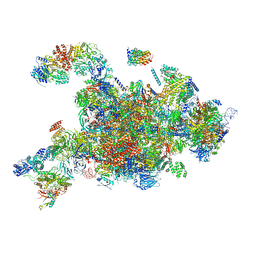 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state a | | Descriptor: | 35S rRNA, 40S ribosomal protein S13-like protein, 40S ribosomal protein S14-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6OAJ
 
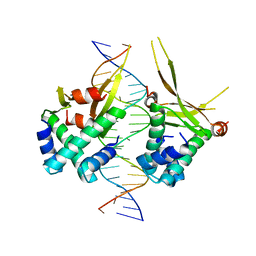 | | HUaE34K 19bp SYM DNA | | Descriptor: | DNA (5'-D(P*CP*GP*GP*TP*TP*CP*AP*AP*TP*TP*GP*GP*CP*AP*CP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*CP*GP*TP*GP*CP*CP*AP*AP*TP*TP*GP*AP*AP*CP*CP*GP*C)-3'), DNA-binding protein HU-alpha | | Authors: | Remesh, S.G, Hammel, M. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.092 Å) | | Cite: | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
7VNY
 
 | | Rba sphaeroides WT RC-LH1 monomer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN B, ... | | Authors: | Bracun, L, Yamagata, A, Liu, L.N, Shirouzu, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis for the assembly and quinone transport mechanisms of the dimeric photosynthetic RC-LH1 supercomplex.
Nat Commun, 13, 2022
|
|
5TJ1
 
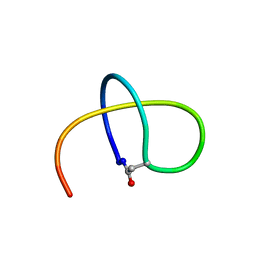 | | Benenodin-1-dC5, state 1 | | Descriptor: | Benenodin-1 | | Authors: | Zong, C, Link, A.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-07-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Lasso Peptide Benenodin-1 Is a Thermally Actuated [1]Rotaxane Switch.
J. Am. Chem. Soc., 139, 2017
|
|
7VP9
 
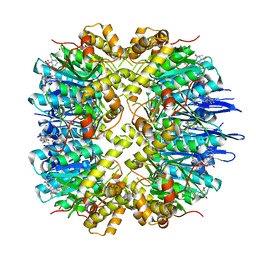 | | Crystal structure of human ClpP in complex with ZG111 | | Descriptor: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Wang, P.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Aberrant human ClpP activation disturbs mitochondrial proteome homeostasis to suppress pancreatic ductal adenocarcinoma.
Cell Chem Biol, 29, 2022
|
|
6S2H
 
 | |
