1Z50
 
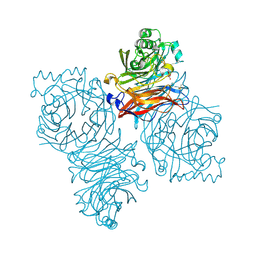 | | Parainfluenza Virus 5 (SV5) Hemagglutinin-Neuraminidase (HN) with ligand DANA (soaked with sialic acid, pH 8.0) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, P, Thompson, T.B, Wurzburg, B.A, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose.
Structure, 13, 2005
|
|
2JH7
 
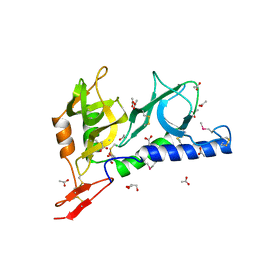 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 6'-sialyl-N-acetyllactosamine | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Blumenschein, T.M.A, Friedrich, N, Childs, R.A, Saouros, S, Carpenter, E.P, Campanero-Rhodes, M.A, Simpson, P, Koutroukides, T, Blackman, M.J, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2007-02-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Atomic Resolution Insight Into Host Cell Recognition by Toxoplasma Gondii.
Embo J., 26, 2007
|
|
1TQ9
 
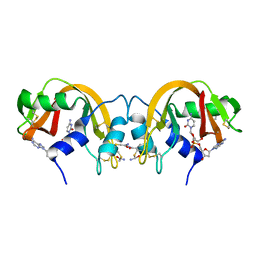 | | Non-covalent swapped dimer of Bovine Seminal Ribonuclease in complex with 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE, Ribonuclease, seminal | | Authors: | Sica, F, Di Fiore, A, Merlino, A, Mazzarella, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Stability of the Non-covalent Swapped Dimer of Bovine Seminal Ribonuclease: AN ENZYME TAILORED TO EVADE RIBONUCLEASE PROTEIN INHIBITOR
J.Biol.Chem., 279, 2004
|
|
2Z8S
 
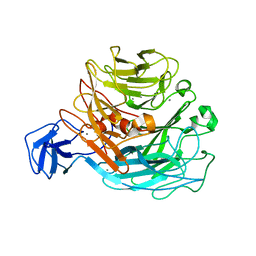 | | Crystal structure of rhamnogalacturonan lyase YesW complexed with digalacturonic acid | | Descriptor: | CALCIUM ION, YesW protein, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Ochiai, A, Itoh, T, Maruyama, Y, Kawamata, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Structural Fold in Polysaccharide Lyases: BACILLUS SUBTILIS FAMILY 11 RHAMNOGALACTURONAN LYASE YesW WITH AN EIGHT-BLADED -PROPELLER
J.Biol.Chem., 282, 2007
|
|
1ZB9
 
 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
2ZE3
 
 | |
2IYU
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with ADP, open LID (conf. A) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IYZ
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with shikimate-3-phosphate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SHIKIMATE KINASE, SHIKIMATE-3-PHOSPHATE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IYR
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-21 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
2IYW
 
 | | Shikimate kinase from Mycobacterium tuberculosis in complex with MgATP, open LID (conf. B) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SHIKIMATE KINASE | | Authors: | Hartmann, M.D, Bourenkov, G.P, Oberschall, A, Strizhov, N, Bartunik, H.D. | | Deposit date: | 2006-07-22 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Phosphoryl Transfer Catalyzed by Shikimate Kinase from Mycobacterium Tuberculosis.
J.Mol.Biol., 364, 2006
|
|
4M63
 
 | | Crystal Structure of a Filament-Like Actin Trimer Bound to the Bacterial Effector VopL | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Zahm, J.A, Rosen, M.K. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.748 Å) | | Cite: | The Bacterial Effector VopL Organizes Actin into Filament-like Structures.
Cell(Cambridge,Mass.), 155, 2013
|
|
2N55
 
 | |
1YQ4
 
 | | Avian respiratory complex ii with 3-nitropropionate and ubiquinone | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 3-NITROPROPANOIC ACID, Coenzyme Q10, ... | | Authors: | Huang, L, Sun, G, Cobessi, D, Wang, A, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
2MYD
 
 | |
2MYB
 
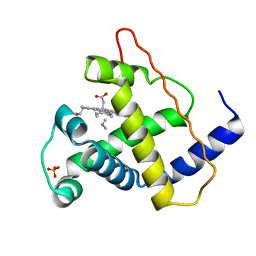 | |
2MYE
 
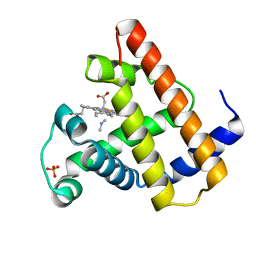 | |
3FIF
 
 | | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A. | | Descriptor: | Uncharacterized ligand, Uncharacterized lipoprotein ygdR | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Rossi, P, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J.C, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-11 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A.
To be Published
|
|
4QB5
 
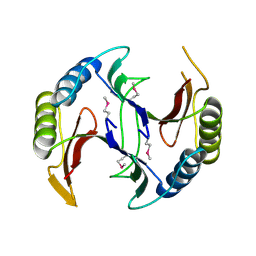 | | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118 | | Descriptor: | 1,2-ETHANEDIOL, Glyoxalase/bleomycin resistance protein/dioxygenase, SULFATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-06 | | Release date: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118
To be Published
|
|
3IDQ
 
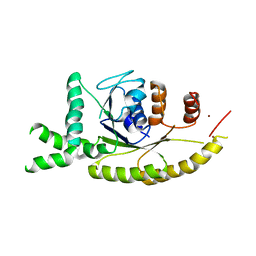 | | Crystal structure of S. cerevisiae Get3 at 3.7 Angstrom resolution | | Descriptor: | ATPase GET3, NICKEL (II) ION, ZINC ION | | Authors: | Suloway, C.J.M, Chartron, J.W, Zaslaver, M, Clemons Jr, W.M. | | Deposit date: | 2009-07-21 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.701 Å) | | Cite: | Model for eukaryotic tail-anchored protein binding based on the structure of Get3
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1YSW
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
2ILU
 
 | |
4M1A
 
 | | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldella termitidis ATCC 33386 | | Descriptor: | Hypothetical protein | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Domain of unknown function (DUF1904) from Sebaldellatermitidis ATCC 33386
To be Published
|
|
2HYR
 
 | | Crystal structure of a complex of griffithsin with maltose | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, MAGNESIUM ION, ... | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-08-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystallographic, thermodynamic, and molecular modeling studies of the mode of binding of oligosaccharides to the potent antiviral protein griffithsin.
Proteins, 67, 2007
|
|
3RQX
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis in complex with P1,P4-Di(adenosine-5') tetraphosphate | | Descriptor: | ADP/ATP-DEPENDENT NAD(P)H-HYDRATE DEHYDRATASE, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
4Q1U
 
 | | Serum paraoxonase-1 by directed evolution with the K192Q mutation | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ben-David, M, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Catalytic stimulation by restrained active-site floppiness-the case of high density lipoprotein-bound serum paraoxonase-1.
J.Mol.Biol., 427, 2015
|
|
