7AVT
 
 | | Crystal structure of SOS1 in complex with compound 7 | | Descriptor: | IMIDAZOLE, Son of sevenless homolog 1, ~{N}-[(1~{R})-1-(3-aminophenyl)ethyl]-6,7-dimethoxy-2-methyl-quinazolin-4-amine | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | One Atom Makes All the Difference: Getting a Foot in the Door between SOS1 and KRAS.
J.Med.Chem., 64, 2021
|
|
7AVI
 
 | | Crystal structure of SOS1 in complex with compound 2 | | Descriptor: | 3-propan-2-yl-~{N}-[(1~{R})-1-(3-sulfamoylphenyl)ethyl]-[1,2]oxazolo[5,4-b]pyridine-5-carboxamide, Son of sevenless homolog 1 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | One Atom Makes All the Difference: Getting a Foot in the Door between SOS1 and KRAS.
J.Med.Chem., 64, 2021
|
|
7AVS
 
 | | Crystal structure of SOS1 in complex with compound 6 | | Descriptor: | 6,7-dimethoxy-2-methyl-~{N}-[(1~{R})-1-[3-(trifluoromethyl)phenyl]ethyl]quinazolin-4-amine, IMIDAZOLE, Son of sevenless homolog 1 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | One Atom Makes All the Difference: Getting a Foot in the Door between SOS1 and KRAS.
J.Med.Chem., 64, 2021
|
|
7AVU
 
 | | Crystal structure of SOS1 in complex with compound 8 | | Descriptor: | IMIDAZOLE, Son of sevenless homolog 1, ~{N}-[(1~{R})-1-[3-azanyl-5-(trifluoromethyl)phenyl]ethyl]-6,7-dimethoxy-2-methyl-quinazolin-4-amine | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | One Atom Makes All the Difference: Getting a Foot in the Door between SOS1 and KRAS.
J.Med.Chem., 64, 2021
|
|
7AVL
 
 | | Crystal structure of SOS1 in complex with compound 4 | | Descriptor: | 6,7-dimethoxy-2-methyl-~{N}-[(1~{R})-1-phenylethyl]quinazolin-4-amine, IMIDAZOLE, Son of sevenless homolog 1 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | One Atom Makes All the Difference: Getting a Foot in the Door between SOS1 and KRAS.
J.Med.Chem., 64, 2021
|
|
7AVV
 
 | | Crystal structure of SOS1 in complex with compound 9 | | Descriptor: | IMIDAZOLE, Son of sevenless homolog 1, ~{N}-[(1~{R})-1-[3-azanyl-5-(trifluoromethyl)phenyl]ethyl]-2-methyl-quinazolin-4-amine | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | One Atom Makes All the Difference: Getting a Foot in the Door between SOS1 and KRAS.
J.Med.Chem., 64, 2021
|
|
8BE4
 
 | |
8BE2
 
 | | Crystal structure of SOS1-Nanobody77 | | Descriptor: | Nanobody 77, SULFATE ION, Son of sevenless homolog 1 | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.897935 Å) | | Cite: | ChILL & DisCO to discover Nanobodies that modulate protein-protein interactions and tune the SOS-Ras nucleotide exchange rate
To Be Published
|
|
8BE5
 
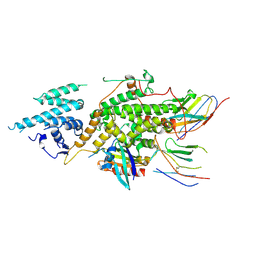 | |
7UKS
 
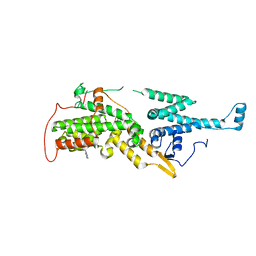 | | Crystal structure of SOS1 with phthalazine inhibitor bound (compound 15) | | Descriptor: | 4-methyl-N-{(1R)-1-[2-methyl-3-(trifluoromethyl)phenyl]ethyl}-7-(piperazin-1-yl)phthalazin-1-amine, Son of sevenless homolog 1 | | Authors: | Gunn, R.J, Lawson, J.D, Ketcham, J.M, Marx, M.A. | | Deposit date: | 2022-04-01 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Design and Discovery of MRTX0902, a Potent, Selective, Brain-Penetrant, and Orally Bioavailable Inhibitor of the SOS1:KRAS Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7UKR
 
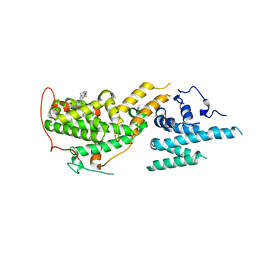 | | Crystal Structure of SOS1 with MRTX0902, a Potent and Selective Inhibitor of the SOS1:KRAS Protein-Protein Interaction | | Descriptor: | 2-methyl-3-[(1R)-1-{[4-methyl-7-(morpholin-4-yl)pyrido[3,4-d]pyridazin-1-yl]amino}ethyl]benzonitrile, Son of sevenless homolog 1 | | Authors: | Gunn, R.J, Lawson, J.D, Ketcham, J.M, Marx, M.A. | | Deposit date: | 2022-04-01 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and Discovery of MRTX0902, a Potent, Selective, Brain-Penetrant, and Orally Bioavailable Inhibitor of the SOS1:KRAS Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
4GBQ
 
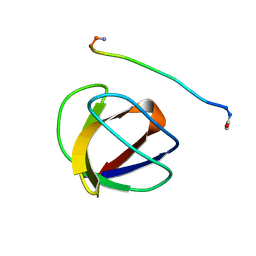 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1S5Z
 
 | | NDP kinase in complex with adenosine phosphonoacetic acid | | Descriptor: | ADENOSINE PHOSPHONOACETIC ACID, Nucleoside diphosphate kinase, cytosolic, ... | | Authors: | Chen, Y, Morera, S, Pasti, C, Angusti, A, Solaroli, N, Veron, M, Janin, J, Manfredini, S, Deville-Bonne, D. | | Deposit date: | 2004-01-22 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Adenosine phosphonoacetic acid is slowly metabolized by NDP kinase.
Med.Chem., 1, 2005
|
|
8T5G
 
 | | SOS2 co-crystal structure with fragment bound (compound 12) | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Son of sevenless homolog 2, ... | | Authors: | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T5R
 
 | | SOS2 crystal structure with fragment bound (compound 13) | | Descriptor: | 4-(aminomethyl)benzene-1-sulfonamide, SULFATE ION, Son of sevenless homolog 2 | | Authors: | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T5M
 
 | | SOS2 crystal structure with fragment bound (compound 14) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1R,2S)-1-hydroxy-2-{[2-(4-hydroxyphenyl)ethyl]amino}propyl]phenol, SULFATE ION, ... | | Authors: | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
2GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
8UH0
 
 | |
8UF2
 
 | | Apo SOS2 crystal structure in P1 space group | | Descriptor: | SULFATE ION, Son of sevenless homolog 2 | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8UC9
 
 | |
1FS4
 
 | | Structures of glycogen phosphorylase-inhibitor complexes and the implications for structure-based drug design | | Descriptor: | 1-DEOXY-1-METHOXYCARBAMIDO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-08 | | Release date: | 2000-10-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
1FU4
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | GLYCOGEN PHOSPHORYLASE, N-[(5S,7R,8S,9S,10R)-8,9,10-trihydroxy-7-(hydroxymethyl)-2,4-dioxo-6-oxa-1,3-diazaspiro[4.5]dec-3-yl]acetamide, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
1FTQ
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | (5S,7R,8S,9S,10R)-3-amino-8,9,10-trihydroxy-7-(hydroxymethyl)-6-oxa-1,3-diazaspiro[4.5]decane-2,4-dione, GLYCOGEN PHOSPHORYLASE, INOSINIC ACID, ... | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
1FTW
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | (5S,7R,8S,9S,10R)-3,8,9,10-tetrahydroxy-7-(hydroxymethyl)-6-oxa-1,3-diazaspiro[4.5]decane-2,4-dione, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
1FU7
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | GLYCOGEN PHOSPHORYLASE, N-(methoxycarbonyl)-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
