1NNI
 
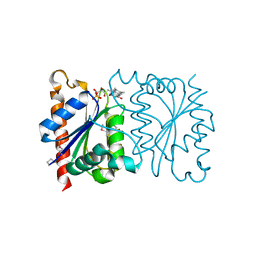 | | Azobenzene Reductase from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein yhda | | Authors: | Cuff, M.E, Kim, Y, Maj, L, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-13 | | Release date: | 2003-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Azobenzene Reductase from Bacillus subtilis
To be Published, 2003
|
|
1JZY
 
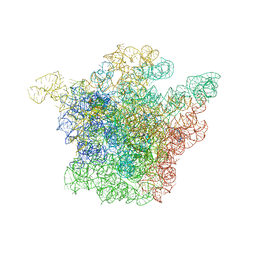 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, ERYTHROMYCIN A, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1K01
 
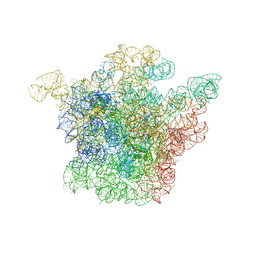 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, CHLORAMPHENICOL, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1JZZ
 
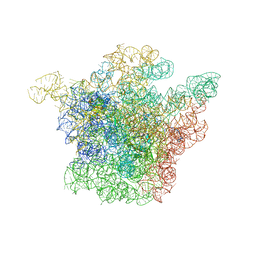 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, MAGNESIUM ION, ROXITHROMYCIN, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
8HJA
 
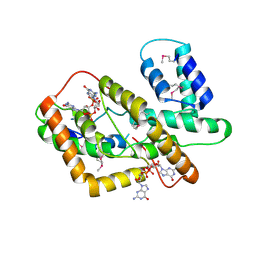 | | The crystal structure of syn_CdgR-(c-di-GMP) from Synechocystis sp. PCC 6803 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), c-di-GMP receptor | | Authors: | Zeng, X, Peng, Y.J. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A c-di-GMP binding effector controls cell size in a cyanobacterium.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1J5A
 
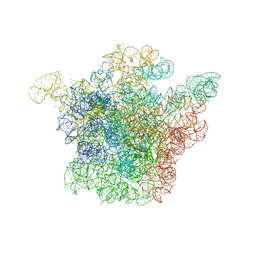 | | STRUCTURAL BASIS FOR THE INTERACTION OF ANTIBIOTICS WITH THE PEPTIDYL TRANSFERASE CENTER IN EUBACTERIA | | Descriptor: | 23S RRNA, CLARITHROMYCIN, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2002-03-06 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1JZX
 
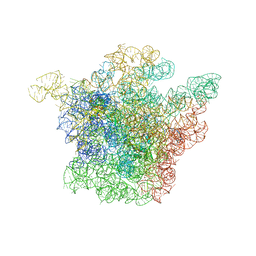 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, CLINDAMYCIN, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1FKA
 
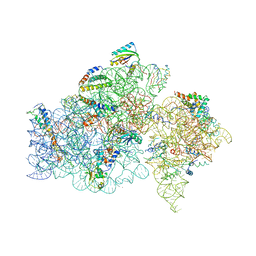 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
8D41
 
 | | Crystal structure of the human COPB2 WD-domain in complex with OICR-6254 | | Descriptor: | (1R,2R,3S,4S)-3-[4-(4-fluorophenyl)piperazine-1-carbonyl]bicyclo[2.2.1]heptane-2-carboxylic acid, Coatomer subunit beta', GLYCEROL, ... | | Authors: | Zeng, H, Saraon, P, Dong, A, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human COPB2 WD-domain in complex with OICR-6254
To Be Published
|
|
8D30
 
 | | Crystal structure of the human COPB2 WD-domains | | Descriptor: | 1,2-ETHANEDIOL, Coatomer subunit beta' | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-31 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human COPB2 WD-domains
To Be Published
|
|
2NLR
 
 | |
1H34
 
 | | Crystal structure of lima bean trypsin inhibitor | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR | | Authors: | Debreczeni, J.E, Bunkoczi, G, Girmann, B, Sheldrick, G.M. | | Deposit date: | 2002-08-21 | | Release date: | 2003-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | In-House Phase Determination of the Lima Bean Trypsin Inhibitor: A Low-Resolution Sulfur-Sad Case
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1HM8
 
 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, GLMU, BOUND TO ACETYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, CALCIUM ION, UDP-N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE | | Authors: | Sulzenbacher, G, Gal, L, Peneff, C, Fassy, F, Bourne, Y. | | Deposit date: | 2000-12-05 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J.Biol.Chem., 276, 2001
|
|
1HM0
 
 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE, GLMU | | Descriptor: | CALCIUM ION, N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | Authors: | Sulzenbacher, G, Gal, L, Peneff, C, Fassy, F, Bourne, Y. | | Deposit date: | 2000-12-04 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J.Biol.Chem., 276, 2001
|
|
1HL8
 
 | |
1HL9
 
 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA ALPHA-FUCOSIDASE IN COMPLEX WITH A MECHANISM BASED INHIBITOR | | Descriptor: | 2-deoxy-2-fluoro-beta-L-fucopyranose, PUTATIVE ALPHA-L-FUCOSIDASE | | Authors: | Sulzenbacher, G, Bignon, C, Bourne, Y, Henrissat, B. | | Deposit date: | 2003-03-14 | | Release date: | 2004-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Thermotoga Maritima {Alpha}-L-Fucosidase: Insights Into the Catalytic Mechanism and the Molecular Basis for Fucosidosis
J.Biol.Chem., 279, 2004
|
|
1HM9
 
 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, GLMU, BOUND TO ACETYL COENZYME A AND UDP-N-ACETYLGLUCOSAMINE | | Descriptor: | ACETYL COENZYME *A, CALCIUM ION, UDP-N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, ... | | Authors: | Sulzenbacher, G, Gal, L, Peneff, C, Fassy, F, Bourne, Y. | | Deposit date: | 2000-12-05 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J.Biol.Chem., 276, 2001
|
|
1GV3
 
 | | The 2.0 Angstrom resolution structure of the catalytic portion of a cyanobacterial membrane-bound manganese superoxide dismutase | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Atzenhofer, W, Regelsberger, G, Jacob, U, Huber, R, Peschek, G.A, Obinger, C. | | Deposit date: | 2002-02-05 | | Release date: | 2002-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0A Resolution Structure of the Catalytic Portion of a Cyanobacterial Membrane-Bound Manganese Superoxide Dismutase
J.Mol.Biol., 321, 2002
|
|
1NWY
 
 | | COMPLEX OF THE LARGE RIBOSOMAL SUBUNIT FROM DEINOCOCCUS RADIODURANS WITH AZITHROMYCIN | | Descriptor: | 23S RIBOSOMAL RRNA, 5S RIBOSOMAL RRNA, AZITHROMYCIN, ... | | Authors: | Schluenzen, F, Harms, J, Franceschi, F, Hansen, H.A.S, Bartels, H, Zarivach, R, Yonath, A. | | Deposit date: | 2003-02-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the antibiotic activity of ketolides and azalides.
Structure, 11, 2003
|
|
1NWX
 
 | | COMPLEX OF THE LARGE RIBOSOMAL SUBUNIT FROM DEINOCOCCUS RADIODURANS WITH ABT-773 | | Descriptor: | 23S RIBOSOMAL RNA, 5S RIBOSOMAL RNA, CETHROMYCIN, ... | | Authors: | Schluenzen, F, Harms, J, Franceschi, F, Hansen, H.A.S, Bartels, H, Zarivach, R, Yonath, A. | | Deposit date: | 2003-02-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the antibiotic activity of ketolides and azalides.
Structure, 11, 2003
|
|
8JCH
 
 | |
8JRC
 
 | |
6VA5
 
 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-4184 | | Descriptor: | 2-(4-methylpiperazin-1-yl)aniline, GLYCEROL, SULFATE ION, ... | | Authors: | Zeng, H, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-16 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-4184
to be published
|
|
5MU5
 
 | | Structure of MAf glycosyltransferase from Magnetospirillum magneticum AMB-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Murat, D, Vincentelli, R, Wu, L.F, Guerardel, Y, Alberto, F. | | Deposit date: | 2017-01-12 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycosylate and move! The glycosyltransferase Maf is involved in bacterial flagella formation.
Environ. Microbiol., 20, 2018
|
|
5AKC
 
 | | MutS in complex with the N-terminal domain of MutL - crystal form 2 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
