5LJ7
 
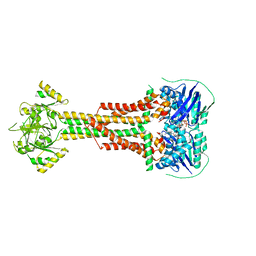 | | Structure of Aggregatibacter actinomycetemcomitans MacB bound to ATP (P21) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Macrolide export ATP-binding/permease protein MacB | | Authors: | Crow, A. | | Deposit date: | 2016-07-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4FZG
 
 | | 20S yeast proteasome in complex with glidobactin | | Descriptor: | Glidobactin, Proteasome component C1, Proteasome component C11, ... | | Authors: | Stein, M, Beck, P, Kaiser, M, Dudler, R, Becker, C.F.W, Groll, M. | | Deposit date: | 2012-07-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1KD6
 
 | | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II | | Descriptor: | EQUINATOXIN II | | Authors: | Hinds, M.G, Zhang, W, Anderluh, G, Hansen, P.E, Norton, R.S. | | Deposit date: | 2001-11-12 | | Release date: | 2002-02-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II: implications for pore formation.
J.Mol.Biol., 315, 2002
|
|
8EOA
 
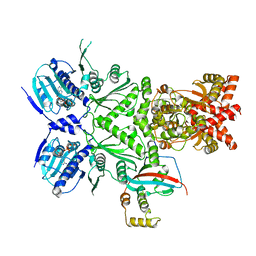 | | Cryo-EM structure of human HSP90B-AIPL1 complex | | Descriptor: | Aryl-hydrocarbon-interacting protein-like 1, Heat shock protein HSP 90-beta, MAGNESIUM ION, ... | | Authors: | Srivastava, D, Artemyev, N.O. | | Deposit date: | 2022-10-02 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Unique interface and dynamics of the complex of HSP90 with a specialized cochaperone AIPL1.
Structure, 31, 2023
|
|
6CO2
 
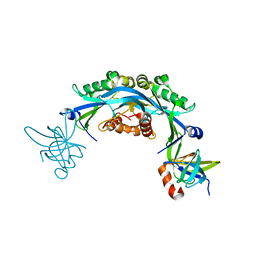 | | Structure of an engineered protein (NUDT16TI) in complex with 53BP1 Tudor domains | | Descriptor: | NUDT16-Tudor-interacting (NUDT16TI), TP53-binding protein 1 | | Authors: | Botuyan, M.V, Thompson, J.R, Cui, G, Mer, G. | | Deposit date: | 2018-03-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6D0L
 
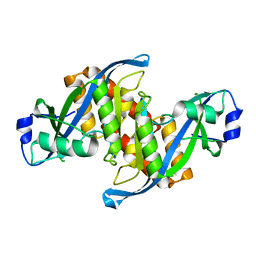 | | Structure of human TIRR | | Descriptor: | Tudor-interacting repair regulator protein | | Authors: | Cui, G, Botuyan, M.V, Mer, G. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4YM4
 
 | | Truncated Human TIFA in complex with its Thr9 phosphorylated N-terminal peptide 1-15 | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Weng, J.H, Wei, T.Y.W, Hsieh, Y.C, Huang, C.C.F, Wu, P.Y.G, Chen, E.S.W, Huang, K.F, Chen, C.J, Tsai, M.D. | | Deposit date: | 2015-03-06 | | Release date: | 2015-10-21 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Uncovering the Mechanism of Forkhead-Associated Domain-Mediated TIFA Oligomerization That Plays a Central Role in Immune Responses.
Biochemistry, 54, 2015
|
|
5APP
 
 | | Actinobacillus actinomycetemcomitans OMP100 residues 133-198 fused to GCN4 adaptors | | Descriptor: | CHLORIDE ION, General control protein GCN4,GENERAL CONTROL PROTEIN GCN4, OUTER MEMBRANE PROTEIN 100,General control protein GCN4 | | Authors: | Hartmann, M.D, Ridderbusch, O, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
6AZA
 
 | |
1BHW
 
 | | LOW TEMPERATURE MIDDLE RESOLUTION STRUCTURE OF XYLOSE ISOMERASE FROM MASC DATA | | Descriptor: | XYLOSE ISOMERASE | | Authors: | Ramin, M, Shepard, W, Fourme, R, Kahn, R. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Multiwavelength anomalous solvent contrast (MASC): derivation of envelope structure-factor amplitudes and comparison with model values.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8JT3
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with amino donor L-Arg | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, ACETATE ION, CrmG, ... | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2023-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Co-crystal structure provides insights on transaminase CrmG recognition amino donor L-Arg.
Biochem.Biophys.Res.Commun., 675, 2023
|
|
5DDU
 
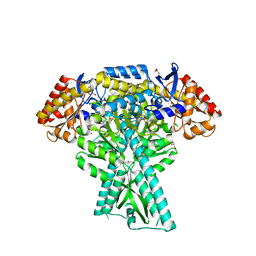 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
4WHN
 
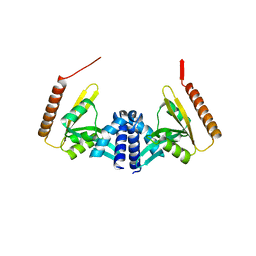 | | Structure of toxin-activating acyltransferase (TAAT) | | Descriptor: | ApxC, CITRIC ACID | | Authors: | Crow, A, Greene, N.P, Hughes, C, Koronakis, V. | | Deposit date: | 2014-09-23 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a bacterial toxin-activating acyltransferase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8QHI
 
 | |
5DDS
 
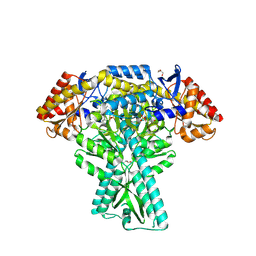 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PLP | | Descriptor: | ACETIC ACID, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
8QHH
 
 | |
5DDW
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with the PMP external aldimine adduct with Caerulomycin M | | Descriptor: | CrmG, GLYCEROL, [5-hydroxy-4-({(E)-[(4-hydroxy-2,2'-bipyridin-6-yl)methylidene]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
1TXQ
 
 | |
3PQS
 
 | |
5UTT
 
 | | SrtA sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Sortase | | Authors: | Osipiuk, J, Ma, X, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cell-to-cell interaction requires optimal positioning of a pilus tip adhesin modulated by gram-positive transpeptidase enzymes.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3QRW
 
 | |
3RI3
 
 | |
3ZVT
 
 | | Unexpected tricovalent binding mode of boronic acids within the active site of a penicillin binding protein | | Descriptor: | 2,6 DIMETHOXYBENZAMIDOBORONIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, Zervosen, A, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2011-07-27 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Unexpected Tricovalent Binding Mode of Boronic Acids within the Active Site of a Penicillin- Binding Protein.
J.Am.Chem.Soc., 133, 2011
|
|
3ZVW
 
 | | Unexpected tricovalent binding mode of boronic acids within the active site of a penicillin binding protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,3-DIMETHYLBUTAN-1-OL, ACETONE, ... | | Authors: | Sauvage, E, Zervosen, A, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2011-07-28 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected Tricovalent Binding Mode of Boronic Acids within the Active Site of a Penicillin- Binding Protein.
J.Am.Chem.Soc., 133, 2011
|
|
2Y4A
 
 | | Unexpected tricovalent binding mode of boronic acids within the active site of a penicillin binding protein | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Sauvage, E, Zervosen, A, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unexpected Tricovalent Binding Mode of Boronic Acids within the Active Site of a Penicillin- Binding Protein.
J.Am.Chem.Soc., 133, 2011
|
|
