6N79
 
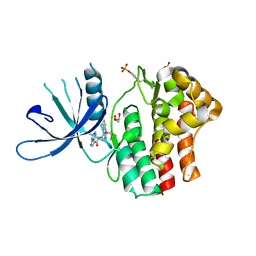 | | Structure of the human JAK1 kinase domain with compound 20 | | Descriptor: | GLYCEROL, N-{5-[5-chloro-2-(difluoromethoxy)phenyl]-1H-pyrazol-4-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
1V5S
 
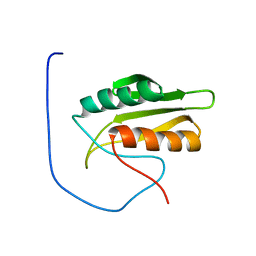 | | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | | Descriptor: | MAP/microtubule affinity-regulating kinase 3 | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3
To be Published
|
|
6N9Q
 
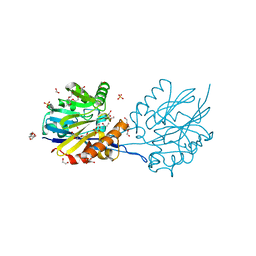 | | Structure of the Quorum Quenching lactonase from Parageobacillus caldoxylosilyticus bind to substrate C4-AHL | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Elias, M. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structural Determinants Accounting for the Broad Substrate Specificity of the Quorum Quenching Lactonase GcL.
Chembiochem, 20, 2019
|
|
2ZJV
 
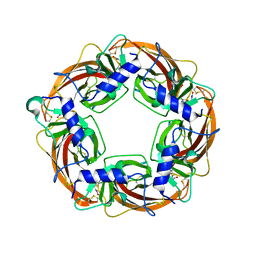 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein (Ls-AChBP) Complexed with Clothianidin | | Descriptor: | 1-[(2-chloro-1,3-thiazol-5-yl)methyl]-3-methyl-2-nitroguanidine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Morimoto, T, Matsuda, K. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Lymnaea stagnalis AChBP in complex with neonicotinoid insecticides imidacloprid and clothianidin
Invert.Neurosci., 8, 2008
|
|
5CLX
 
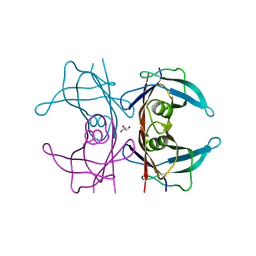 | | X-ray structure of perdeuterated TTR mutant - S52P at 1.28A resolution | | Descriptor: | GLYCEROL, Transthyretin | | Authors: | Yee, A.W, Moulin, M, Mossou, E, Haertlein, M, Mitchell, E.P, Cooper, J.B, Forsyth, V.T. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | X-ray structure of perdeuterated TTR mutant - S52P at 1.28A resolution
To Be Published
|
|
4KSG
 
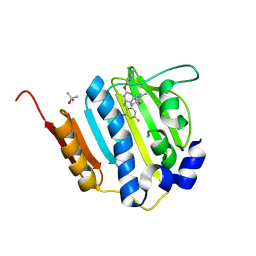 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor (4-[(1S,5R,6R)-6-AMINO-1-METHYL-3-AZABICYCLO[3.2.0]HEPT-3-YL]-6-FLUORO-N-METHYL-2-[(2-METHYLPYRIMIDIN-5-YL)OXY]-9H-PYRIMIDO[4,5-B]INDOL-8-AMINE) | | Descriptor: | 4-[(1S,5R,6R)-6-amino-1-methyl-3-azabicyclo[3.2.0]hept-3-yl]-6-fluoro-N-methyl-2-[(2-methylpyrimidin-5-yl)oxy]-9H-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | Authors: | Bensen, D.C, Akers-Rodriguez, S, Tari, L.W. | | Deposit date: | 2013-05-17 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A new class of type iia topoisomerase inhibitors with
broad-spectrum antibacterial activity
To be Published
|
|
3W05
 
 | | Crystal structure of Oryza sativa DWARF14 (D14) in complex with PMSF | | Descriptor: | 1,2-ETHANEDIOL, Dwarf 88 esterase, phenylmethanesulfonic acid | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
4QJX
 
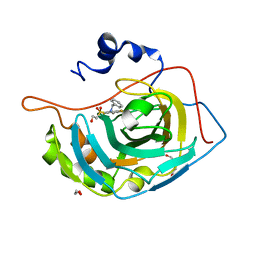 | | Crystal structure of human carbonic anhydrase isozyme XIII with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 13, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-06-05 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functionalization of Fluorinated Benzenesulfonamides and Their Inhibitory Properties toward Carbonic Anhydrases
Chemmedchem, 10, 2015
|
|
6MX9
 
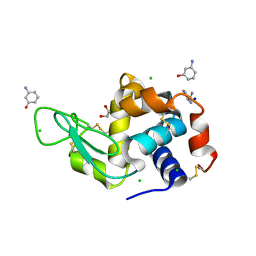 | | Lysozyme bound to 3-Aminophenol | | Descriptor: | 1,2-ETHANEDIOL, 3-aminophenol, BENZAMIDINE, ... | | Authors: | Blackburn, A, Partowmah, S.H, Brennan, H.M, Mestizo, K.E, Stivala, C.D, Petreczky, J, Perez, A, Horn, A, McSweeney, S. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A simple technique to improve microcrystals using gel exclusion of nucleation inducing elements
To Be Published
|
|
5VAU
 
 | |
6MWR
 
 | | Recognition of MHC-like molecule | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Le Nours, J, Rossjohn, J. | | Deposit date: | 2018-10-30 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A class of gamma delta T cell receptors recognize the underside of the antigen-presenting molecule MR1.
Science, 366, 2019
|
|
5XND
 
 | |
5VE7
 
 | |
5XC1
 
 | | Crystal structure of the complex of an aromatic mutant (W6A) of an alkali thermostable GH10 Xylanase from Bacillus sp. NG-27 with S-1,2-Propanediol | | Descriptor: | Beta-xylanase, MAGNESIUM ION, S-1,2-PROPANEDIOL, ... | | Authors: | Bansia, H, Mahanta, P, Ramakumar, S. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Small Glycols Discover Cryptic Pockets on Proteins for Fragment-Based Approaches.
J.Chem.Inf.Model., 2021
|
|
6DGK
 
 | |
6DHI
 
 | |
5FCW
 
 | | HDAC8 Complexed with a Hydroxamic Acid | | Descriptor: | 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Cole, K.E, Perry, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structure of 'linkerless' hydroxamic acid inhibitor-HDAC8 complex confirms the formation of an isoform-specific subpocket.
J.Struct.Biol., 195, 2016
|
|
1OM0
 
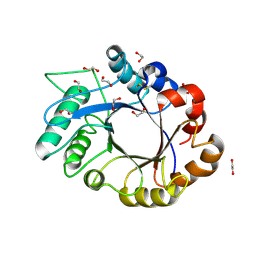 | | crystal structure of xylanase inhibitor protein (XIP-I) from wheat | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Xylanase Inhibitor Protein I | | Authors: | Payan, F, Flatman, R, Porciero, S, Williamson, G, Juge, N, Roussel, A. | | Deposit date: | 2003-02-24 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of xylanase inhibitor protein I (XIP-I), a proteinaceous xylanase inhibitor from wheat (Triticum aestivum, var. Soisson).
Biochem.J., 372, 2003
|
|
5C55
 
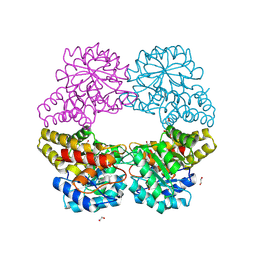 | |
4LXQ
 
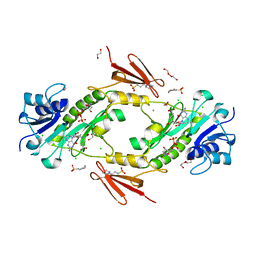 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTdp and 5-N-Formyl-THF | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4CH4
 
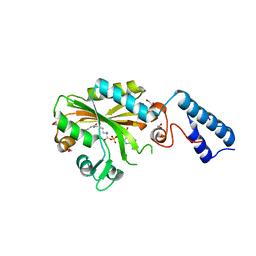 | | Structure of pyrrolysyl-tRNA synthetase in complex with adenylated crotonyl lysine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[({(2R)-2-amino-6-[(2E)-but-2-enoylamino]hexanoyl}oxy)phosphinato]adenosine, MAGNESIUM ION, ... | | Authors: | Fluegel, V, Vrabel, M, Schneider, S. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for the Site-Specific Incorporation of Lysine Derivatives Into Proteins.
Plos One, 9, 2014
|
|
2VYO
 
 | | Chitin deacetylase family member from Encephalitozoon cuniculi | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Urch, J.E, Hurtado-Guerrero, R, Texier, C, Van Aalten, D.M.F. | | Deposit date: | 2008-07-25 | | Release date: | 2008-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of a Putative Polysaccharide Deacetylase of the Human Parasite Encephalitozoon Cuniculi.
Protein Sci., 18, 2009
|
|
6AA1
 
 | | Crystal structure of putative amino acid binding periplasmic ABC transporter protein from Candidatus Liberibacter asiaticus bound with citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, P, Kesari, P, Ghosh, D.K, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-07-16 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of a putative periplasmic cystine-binding protein from Candidatus Liberibacter asiaticus: insights into an adapted mechanism of ligand binding.
Febs J., 286, 2019
|
|
2W22
 
 | | Activation Mechanism of Bacterial Thermoalkalophilic Lipases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, CALCIUM ION, ... | | Authors: | Carrasco-Lopez, C, Godoy, C, De Las Rivas, B, Fernandez-Lorente, G, Palomo, J.M, Guisan, J.M, Fernandez-Lafuente, R, Hermoso, J.A. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of bacterial thermoalkalophilic lipases is spurred by dramatic structural rearrangements.
J. Biol. Chem., 284, 2009
|
|
5CG5
 
 | | Neutron crystal structure of human farnesyl pyrophosphate synthase in complex with risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Yokoyama, T, Mizuguchi, M, Ostermann, A, Kusaka, K, Niimura, N, Schrader, T.E, Tanaka, I. | | Deposit date: | 2015-07-09 | | Release date: | 2015-10-14 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.402 Å), X-RAY DIFFRACTION | | Cite: | Protonation State and Hydration of Bisphosphonate Bound to Farnesyl Pyrophosphate Synthase
J.Med.Chem., 58, 2015
|
|
