8UQB
 
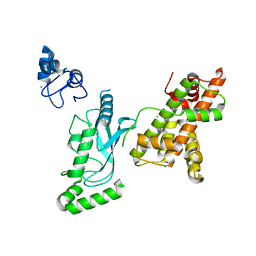 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 1) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UPF
 
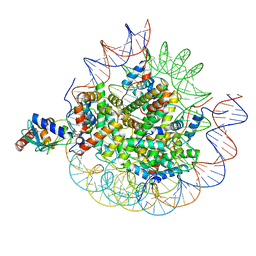 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168-UbcH5c | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQ9
 
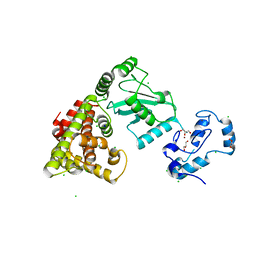 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 4-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
5NB5
 
 | |
8UQ8
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 2-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQD
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (condition 2. RING not modeled in density) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.893 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
2BR7
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, Van Nierop, P, Van Elk, R, Van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha- Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
8UQC
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 2) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
2BR8
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with an alpha-conotoxin PnIA variant | | Descriptor: | ALPHA-CONOTOXIN PNIA, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, van Nierop, P, van Elk, R, van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha-Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
9FBX
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 5-(1-benzyl-4-chloro-1H-imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5-(1-benzyl-4-chloro-1H-imidazol-2-yl)-1,3-dimethylpyridin-2(1H)-one, ... | | Authors: | Chung, C. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structure- and Property-Based Optimization of Efficient Pan-Bromodomain and Extra Terminal Inhibitors to Identify Oral and Intravenous Candidate I-BET787.
J.Med.Chem., 67, 2024
|
|
1XKU
 
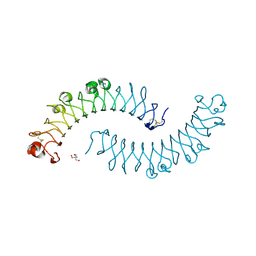 | | Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Decorin | | Authors: | Scott, P.G, McEwan, P.A, Dodd, C.M, Bergmann, E.M, Bishop, P.N, Bella, J. | | Deposit date: | 2004-09-29 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5ZDM
 
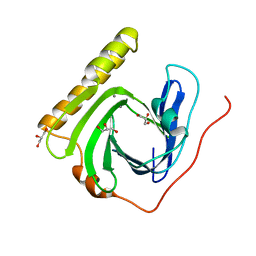 | | The ligand-free structure of FomD | | Descriptor: | CALCIUM ION, FomD, GLYCEROL | | Authors: | Sato, S, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2018-02-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Biochemical and Structural Analysis of FomD That Catalyzes the Hydrolysis of Cytidylyl ( S)-2-Hydroxypropylphosphonate in Fosfomycin Biosynthesis.
Biochemistry, 57, 2018
|
|
3A04
 
 | |
7KOO
 
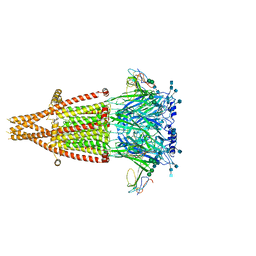 | | Alpha-7 nicotinic acetylcholine receptor bound to alpha-bungarotoxin in a resting state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-bungarotoxin isoform V31, ... | | Authors: | Noviello, C.M, Hibbs, R.E, Gharpure, A, Mukhtasimova, N, Cabuco, R, Baxter, L, Borek, D, Sine, S. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and gating mechanism of the alpha 7 nicotinic acetylcholine receptor.
Cell, 184, 2021
|
|
7KOQ
 
 | | Alpha-7 nicotinic acetylcholine receptor bound to epibatidine in a desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Noviello, C.M, Hibbs, R.E, Gharpure, A, Mukhtasimova, N, Cabuco, R, Baxter, L, Borek, D, Sine, S. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and gating mechanism of the alpha 7 nicotinic acetylcholine receptor.
Cell, 184, 2021
|
|
2BHQ
 
 | | Crystal Analysis of 1-Pyrroline-5-Carboxylate Dehydrogenase from Thermus with bound product glutamate. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Tahirov, T.H. | | Deposit date: | 2005-01-16 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
5FO4
 
 | | Crystal structure of the P.falciparum cytosolic leucyl-tRNA synthetase editing domain (space group P1) | | Descriptor: | LEUCYL TRNA SYNTHASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FOC
 
 | | Crystal structure of the P.falciparum cytosolic leucyl-tRNA synthetase editing domain (space group P21) | | Descriptor: | LEUCYL-TRNA SYNTHETASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FOD
 
 | | Crystal structure of the P.falciparum cytosolic leucyl-tRNA synthetase editing domain (space group P1) containing deletions of insertions 1 and 3 | | Descriptor: | 1,2-ETHANEDIOL, LEUCYL-TRNA SYNTHETASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
7TZB
 
 | | Crystal structure of the human mitochondrial seryl-tRNA synthetase (mt SerRS) bound with a seryl-adenylate analogue | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Serine--tRNA ligase, mitochondrial | | Authors: | Kuhle, B, Hirschi, M, Doerfel, L, Lander, G, Schimmel, P. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
5FOF
 
 | | Crystal structure of the P.knowlesi cytosolic leucyl-tRNA synthetase editing domain | | Descriptor: | LEUCYL-TRNA SYNTHETASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5NKN
 
 | | Crystal structure of an Anticalin-colchicine complex | | Descriptor: | N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide, Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A, Barkovskiy, M. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An engineered lipocalin that tightly complexes the plant poison colchicine for use as antidote and in bioanalytical applications.
Biol. Chem., 400, 2019
|
|
7KK7
 
 | | crystal structure of ligand-free PLEKHA7 PH domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
2BJK
 
 | | Crystal Analysis of 1-Pyrroline-5-Carboxylate Dehydrogenase from Thermus with bound NAD and citrate. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Tahirov, T.H. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
5UU9
 
 | |
