3TP6
 
 | |
3TPA
 
 | |
3HHK
 
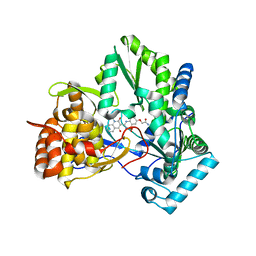 | | HCV NS5b polymerase complex with a substituted benzothiadizine | | Descriptor: | 2-({(3R)-3-[(3S)-1-(3-methylbutyl)-2,4-dioxo-1,2,3,4-tetrahydroquinolin-3-yl]-1,1-dioxido-3,4-dihydro-2H-1,2,4-benzothiadiazin-7-yl}oxy)acetamide, HCV NS5 polymerase | | Authors: | Concha, N.O, Singh, O. | | Deposit date: | 2009-05-15 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted benzothiadizine inhibitors of Hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5WJF
 
 | | Crystal structure of murine 4-1BB from HEK293T cells in P21212 space group | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Zajonc, D.M, Doukov, T, Bitra, A. | | Deposit date: | 2017-07-21 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of murine 4-1BB and its interaction with 4-1BBL support a role for galectin-9 in 4-1BB signaling.
J. Biol. Chem., 293, 2018
|
|
3TLE
 
 | |
3HI5
 
 | | Crystal structure of Fab fragment of AL-57 | | Descriptor: | Heavy chain of Fab fragment of AL-57 against alpha L I domain, light chain of Fab fragment of AL-57 against alpha L I domain | | Authors: | Zhang, H, Wang, J. | | Deposit date: | 2009-05-18 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of activation-dependent binding of ligand-mimetic antibody AL-57 to integrin LFA-1.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4V3E
 
 | | The CIDRa domain from IT4var07 PfEMP1 bound to endothelial protein C receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOTHELIAL PROTEIN C RECEPTOR, ... | | Authors: | Lau, C.K.Y, Turner, L, Jespersen, J.S, Lowe, E.D, Petersen, B, Wang, C.W, Petersen, J.E.V, Lusingu, J, Theander, T.G, Lavstsen, T, Higgins, M.K. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Conservation Despite Huge Sequence Diversity Allows Epcr Binding by the Pfemp1 Family Implicated in Severe Childhood Malaria.
Cell Host Microbe., 17, 2015
|
|
3TPQ
 
 | |
3TR0
 
 | | Structure of guanylate kinase (gmk) from Coxiella burnetii | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, SULFATE ION | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3HGG
 
 | | Crystal Structure of CmeR Bound to Cholic Acid | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-13 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: Crystal structures of CmeR-bile acid complexes
To be Published
|
|
3TRE
 
 | | Structure of a translation elongation factor P (efp) from Coxiella burnetii | | Descriptor: | Elongation factor P | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TM2
 
 | | Crystal structure of mature ThnT with a covalently bound product mimic | | Descriptor: | (2R)-N-(4-chloro-3-oxobutyl)-2,4-dihydroxy-3,3-dimethylbutanamide, cysteine transferase | | Authors: | Schildbach, J.F, Wright, N.T, Buller, A.R. | | Deposit date: | 2011-08-30 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Autoproteolytic Activation of ThnT Results in Structural Reorganization Necessary for Substrate Binding and Catalysis.
J.Mol.Biol., 422, 2012
|
|
3HIQ
 
 | |
3TN5
 
 | | Crystal structure of GkaP mutant Y99L from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, PHOSPHATE ION, Phosphotriesterase | | Authors: | An, J, Zhang, Z, Zhang, Y, Feng, Y, Wu, G. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering a thermostable lactonase for enhanced phosphotriesterase activity against organophosphate pesticides
to be published
|
|
4UZ6
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM - CRYSTAL FORM V - SOS COMPLEX - 1.9A | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Nature, 519, 2015
|
|
3TNW
 
 | | Structure of CDK2/cyclin A in complex with CAN508 | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Baumli, S, Hole, A.J, Endicott, J.A. | | Deposit date: | 2011-09-02 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
3HH1
 
 | | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Tetrapyrrole methylase family protein | | Authors: | Cuff, M.E, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS.
TO BE PUBLISHED
|
|
3HGS
 
 | | Crystal structure of tomato OPR3 in complex with pHB | | Descriptor: | 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Clausen, T, Breithaupt, C. | | Deposit date: | 2009-05-14 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate specificity of plant 12-oxophytodienoate reductases.
J.Mol.Biol., 392, 2009
|
|
3HH7
 
 | | Structural and Functional Characterization of a Novel Homodimeric Three-finger Neurotoxin from the Venom of Ophiophagus hannah (King Cobra) | | Descriptor: | Muscarinic toxin-like protein 3 homolog | | Authors: | Roy, A, Zhou, X, Chong, M.Z, D'hoedt, D, Foo, C.S, Rajagopalan, N, Nirthanan, S, Bertrand, D, Sivaraman, J, Kini, R.M. | | Deposit date: | 2009-05-15 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Characterization of a Novel Homodimeric Three-finger Neurotoxin from the Venom of Ophiophagus hannah (King Cobra)
J.Biol.Chem., 285, 2010
|
|
7M8L
 
 | | EBOV GP bound to rEBOV-442 and rEBOV-515 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Virion spike glycoprotein 1, ... | | Authors: | Murin, C.D, Ward, A.B. | | Deposit date: | 2021-03-30 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Pan-ebolavirus protective therapy by two multifunctional human antibodies.
Cell, 184, 2021
|
|
3HDF
 
 | |
3TV2
 
 | |
3HHS
 
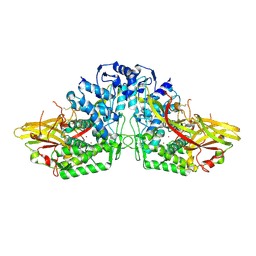 | | Crystal Structure of Manduca sexta prophenoloxidase | | Descriptor: | COPPER (II) ION, Phenoloxidase subunit 1, Phenoloxidase subunit 2 | | Authors: | Li, Y, Wang, Y, Jiang, H, Deng, J. | | Deposit date: | 2009-05-17 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HES
 
 | | Human prion protein variant F198S with M129 | | Descriptor: | CADMIUM ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-10 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3TQU
 
 | | Structure of a HAM1 protein from Coxiella burnetii | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Non-canonical purine NTP pyrophosphatase | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
