8EXC
 
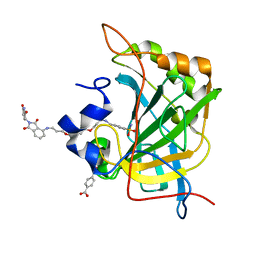 | | Human Carbonic Anhydrase II bound tert-butyl (3-(4-(3-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)propoxy)butoxy)propyl)carbamate | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, N-(3-{4-[3-({2-[(3R)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}amino)propoxy]butoxy}propyl)-4-sulfamoylbenzamide, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
4FCR
 
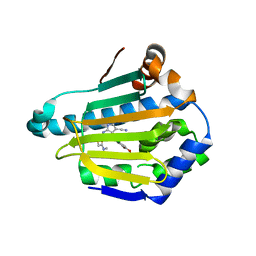 | | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization | | Descriptor: | 2-{[4-(2-chloro-4,5-dimethoxyphenyl)-5-cyano-7H-pyrrolo[2,3-d]pyrimidin-2-yl]sulfanyl}-N,N-dimethylacetamide, Heat shock protein HSP 90-alpha | | Authors: | Davies, N.G, Browne, H, Davis, B, Foloppe, N, Geoffrey, S, Gibbons, B, Hart, T, Drysdale, M.J, Mansell, H, Massey, A, Matassova, N, Moore, J.D, Murray, J, Pratt, R, Ray, S, Roughley, S.D, Jensen, M.R, Schoepfer, J, Scriven, K, Simmonite, H, Stokes, S, Surgenor, A, Webb, P, Wright, L, Brough, P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization.
Bioorg.Med.Chem., 20, 2012
|
|
2FTB
 
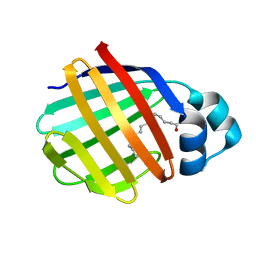 | | Crystal structure of axolotl (Ambystoma mexicanum) liver bile acid-binding protein bound to oleic acid | | Descriptor: | Fatty acid-binding protein 2, liver, OLEIC ACID | | Authors: | Capaldi, S, Guariento, M, Perduca, M, Di Pietro, S.M, Santome, J.A, Monaco, H.L. | | Deposit date: | 2006-01-24 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of axolotl (Ambystoma mexicanum) liver bile acid-binding protein bound to cholic and oleic acid
Proteins, 64, 2006
|
|
8EYL
 
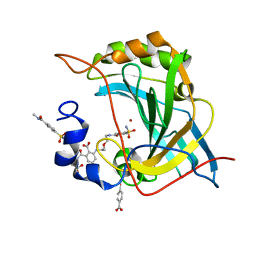 | | Human Carbonic Anhydrase II with Tert-butyl (2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethyl)carbamate | | Descriptor: | 2-[[(2~{R})-1-azanyl-5-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]carbamoyl]-6-[2-[2-[(4-sulfamoylphenyl)carbonylamino]ethoxy]ethylamino]benzoic acid, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-27 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
1R5Y
 
 | | Crystal Structure of TGT in complex with 2,6-Diamino-3H-Quinazolin-4-one Crystallized at PH 5.5 | | Descriptor: | 2,6-DIAMINO-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Garcia, G.A, Stubbs, M.T, Klebe, G. | | Deposit date: | 2003-10-13 | | Release date: | 2004-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1B4T
 
 | | H48C YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 1999-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
4FF6
 
 | | Mycobacterium tuberculosis DprE1 in complex with CT325 - monoclinic crystal form | | Descriptor: | 3-(hydroxyamino)-N-[(1R)-1-phenylethyl]-5-(trifluoromethyl)benzamide, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Batt, S.M, Besra, G.S, Futterer, K. | | Deposit date: | 2012-05-31 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3WK3
 
 | |
3FST
 
 | |
2WMQ
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | N-(4-OXO-5,6,7,8-TETRAHYDRO-4H-[1,3]THIAZOLO[5,4-C]AZEPIN-2-YL)ACETAMIDE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
4QH1
 
 | | The GLIC pentameric Ligand-Gated Ion Channel (wild-type) in complex with bromoacetate | | Descriptor: | BROMIDE ION, Proton-gated ion channel, SODIUM ION, ... | | Authors: | Fourati, Z, Delarue, M, Sauguet, L. | | Deposit date: | 2014-05-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural characterization of potential modulation sites in the extracellular domain of the prokaryotic pentameric proton-gated ion channel GLIC
Acta Crystallogr.,Sect.D, 2015
|
|
1X9F
 
 | | Hemoglobin Dodecamer from Lumbricus Erythrocruorin | | Descriptor: | CARBON MONOXIDE, Globin II, extracellular, ... | | Authors: | Strand, K, Knapp, J.E, Bhyravbhatla, B, Royer Jr, W.E. | | Deposit date: | 2004-08-20 | | Release date: | 2004-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the hemoglobin dodecamer from lumbricus erythrocruorin: allosteric core of giant annelid respiratory complexes
J.Mol.Biol., 344, 2004
|
|
2JKW
 
 | | Pseudoazurin M16F | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Yanagisawa, S, Crowley, P.B, Firbank, S.J, Lawler, A.T, Hunter, D.M, McFarlane, W, Li, C, Kohzuma, T, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pi-Interaction Tuning of the Active Site Properties of Metalloproteins.
J.Am.Chem.Soc., 130, 2008
|
|
7VEE
 
 | | The ligand-free structure of GfsA KSQ-AT didomain | | Descriptor: | GLYCEROL, Polyketide synthase | | Authors: | Chisuga, T, Miyanaga, A, Nagai, A, Kudo, F, Eguchi, T. | | Deposit date: | 2021-09-08 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insight into the Reaction Mechanism of Ketosynthase-Like Decarboxylase in a Loading Module of Modular Polyketide Synthases.
Acs Chem.Biol., 17, 2022
|
|
3WJZ
 
 | | Orotidine 5'-monophosphate decarboxylase D75N mutant from M. thermoautotrophicus complexed with 6-amino-UMP | | Descriptor: | 6-AMINOURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fujihashi, M, Pai, E.F, Miki, K. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate distortion contributes to the catalysis of orotidine 5'-monophosphate decarboxylase.
J.Am.Chem.Soc., 135, 2013
|
|
4DHI
 
 | | Structure of C. elegans OTUB1 bound to human UBC13 | | Descriptor: | Ubiquitin thioesterase otubain-like, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
3S1I
 
 | | Crystal structure of oxygen-bound hell's gate globin I | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
9HFU
 
 | | Caprin1 peptide bound to SPOP MATH domain | | Descriptor: | Caprin-1, SODIUM ION, Speckle type BTB/POZ protein | | Authors: | Makhlouf, L, Zeqiraj, E. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sequence rules for a long SPOP-binding degron required for protein ubiquitylation.
Biochem.J., 482, 2025
|
|
9HFW
 
 | | SRC-3 (NCoA-3) peptide bound to SPOP MATH domain | | Descriptor: | Nuclear receptor coactivator 3, Speckle type BTB/POZ protein | | Authors: | Makhlouf, L, Zeqiraj, E. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sequence rules for a long SPOP-binding degron required for protein ubiquitylation.
Biochem.J., 482, 2025
|
|
9HGG
 
 | | SETD2 peptide bound to SPOP MATH domain | | Descriptor: | Histone-lysine N-methyltransferase SETD2, MAGNESIUM ION, Speckle-type POZ protein | | Authors: | Makhlouf, L, Zeqiraj, E. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sequence rules for a long SPOP-binding degron required for protein ubiquitylation.
Biochem.J., 482, 2025
|
|
4XID
 
 | | AntpHD with 15bp DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2015-01-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
9HGH
 
 | | MyD88 peptide_1 bound to SPOP MATH domain | | Descriptor: | Myeloid differentiation primary response protein MyD88, Speckle type BTB/POZ protein | | Authors: | Makhlouf, L, Zeqiraj, E. | | Deposit date: | 2024-11-19 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sequence rules for a long SPOP-binding degron required for protein ubiquitylation.
Biochem.J., 482, 2025
|
|
4DHJ
 
 | | The structure of a ceOTUB1 ubiquitin aldehyde UBC13~Ub complex | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
4ZDM
 
 | | Pleurobrachia bachei iGluR3 LBD Glycine Complex | | Descriptor: | GLYCINE, Glutamate receptor kainate-like protein, SODIUM ION, ... | | Authors: | Grey, R.J, Mayer, M.L. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Glycine activated ion channel subunits encoded by ctenophore glutamate receptor genes.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3OKL
 
 | | Crystal structure of S25-39 in complex with Kdo(2.8)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
