2W3Z
 
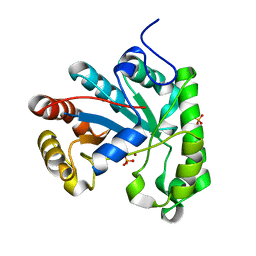 | | Structure of a Streptococcus mutans CE4 esterase | | Descriptor: | PHOSPHATE ION, PUTATIVE DEACETYLASE, ZINC ION | | Authors: | Deng, D.M, Urch, J.E, ten Cate, J.M, Rao, V.A, van Aalten, D.M.F, Crielaard, W. | | Deposit date: | 2008-11-17 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Streptococcus Mutans Smu.623C Codes for a Functional, Metal Dependent Polysaccharide Deacetylase that Modulates Interactions with Salivary Agglutinin.
J.Bacteriol., 191, 2009
|
|
3QLH
 
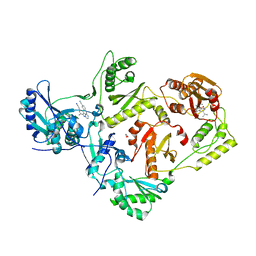 | | HIV-1 Reverse Transcriptase in Complex with Manicol at the RNase H Active Site and TMC278 (Rilpivirine) at the NNRTI Binding Pocket | | Descriptor: | (2S)-5,7-dihydroxy-9-methyl-2-(prop-1-en-2-yl)-1,2,3,4-tetrahydro-6H-benzo[7]annulen-6-one, 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Himmel, D.M, Wojtak, K, Bauman, J.D, Arnold, E. | | Deposit date: | 2011-02-02 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis, activity, and structural analysis of novel alpha-hydroxytropolone inhibitors of human immunodeficiency virus reverse transcriptase-associated ribonuclease H.
J.Med.Chem., 54, 2011
|
|
3R6P
 
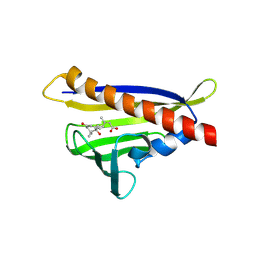 | | Crystal structure of abscisic acid-bound PYL10 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL10 | | Authors: | Sun, D.M, Wu, M.H, Wang, H.P, Zang, J.Y, Tian, C.L. | | Deposit date: | 2011-03-22 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of abscisic acid-bound PYL10
To be Published
|
|
1TYC
 
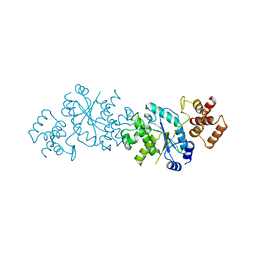 | |
1TYB
 
 | |
1TYA
 
 | |
1TYD
 
 | |
3O2U
 
 | | S. cerevisiae Ubc12 | | Descriptor: | GLYCEROL, NEDD8-conjugating enzyme UBC12 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
3OEH
 
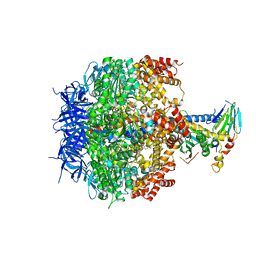 | | Structure of four mutant forms of yeast F1 ATPase: beta-V279F | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
3O2P
 
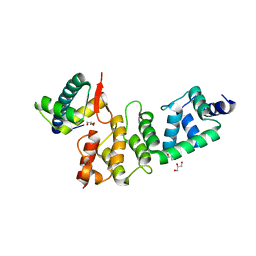 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1, GLYCEROL | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
3OE7
 
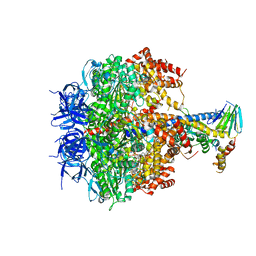 | | Structure of four mutant forms of yeast f1 ATPase: gamma-I270T | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
3O8X
 
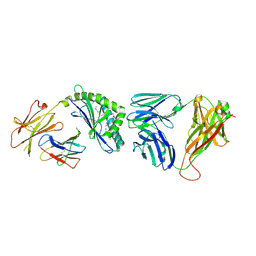 | | Recognition of Glycolipid Antigen by iNKT Cell TCR | | Descriptor: | (2S,3R)-3-HYDROXY-2-(TETRADECANOYLAMINO)OCTADECYL ALPHA-D-GALACTOPYRANOSIDURONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Li, Y. | | Deposit date: | 2010-08-03 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The V alpha 14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode.
J.Exp.Med., 207, 2010
|
|
3O9W
 
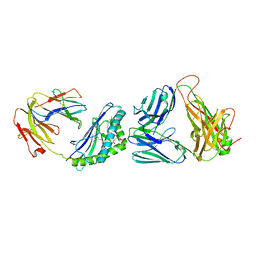 | | Recognition of a Glycolipid Antigen by the iNKT Cell TCR | | Descriptor: | (2S)-3-(alpha-D-galactopyranosyloxy)-2-(hexadecanoyloxy)propyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Li, Y. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The V alpha 14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode.
J.Exp.Med., 207, 2010
|
|
3OMJ
 
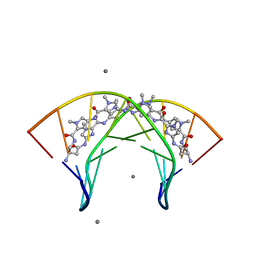 | | Structural Basis for Cyclic Py-Im Polyamide Allosteric Inhibition of Nuclear Receptor Binding | | Descriptor: | (23R,52R)-23,52-diamino-5,11,17,28,34,40,46,57-octamethyl-2,5,8,11,14,17,20,25,28,31,34,37,40,43,46,49,54,57,60,64-icosaazanonacyclo[54.2.1.1~4,7~.1~10,13~.1~16,19~.1~27,30~.1~33,36~.1~39,42~.1~45,48~]hexahexaconta-1(58),4(66),6,10(65),12,16(64),18,27(63),29,33(62),35,39(61),41,45(60),47,56(59)-hexadecaene-3,9,15,21,26,32,38,44,50,55-decone, 5'-D(*CP*(C38)P*AP*GP*TP*AP*CP*TP*GP*G)-3', CALCIUM ION | | Authors: | Chenoweth, D.M. | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for cyclic py-im polyamide allosteric inhibition of nuclear receptor binding.
J.Am.Chem.Soc., 132, 2010
|
|
3OJD
 
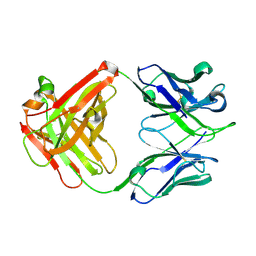 | |
3P0G
 
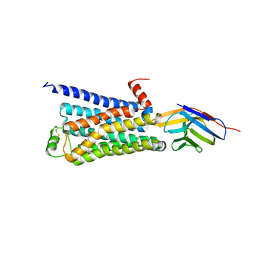 | | Structure of a nanobody-stabilized active state of the beta2 adrenoceptor | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Rasmussen, S.G.F, Choi, H.-J, Fung, J.J, Pardon, E, Casarosa, P, Chae, P.S, DeVree, B.T, Rosenbaum, D.M, Thian, F.S, Kobilka, T.S, Schnapp, A, Konetzki, I, Sunahara, R.K, Gellman, S.H, Pautsch, A, Steyaert, J, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2010-09-28 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a nanobody-stabilized active state of the b2 adrenoceptor
Nature, 469, 2011
|
|
3OR7
 
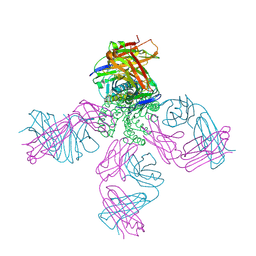 | | On the structural basis of modal gating behavior in K+channels - E71I | | Descriptor: | POTASSIUM ION, Voltage-gated potassium channel, antibody fab fragment heavy chain, ... | | Authors: | Chakrapani, S, Cordero-Morales, J.F, Jogini, V, Pan, A.C, Cortes, D.M, Roux, B, Perozo, E. | | Deposit date: | 2010-09-06 | | Release date: | 2011-01-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the structural basis of modal gating behavior in K(+) channels.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3PWY
 
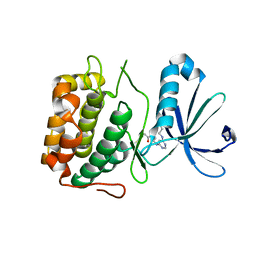 | | Crystal structure of an extender (SPD28345)-modified human PDK1 complex 2 | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, N-[2-({6-[(2-sulfanylethyl)amino]pyrimidin-4-yl}amino)ethyl]propanamide | | Authors: | Elling, R.A, Penny, D.M, Simmons, R.L, Erlanson, D.A, Romanowski, M.J. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of a potent and highly selective PDK1 inhibitor via fragment-based drug discovery.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PSC
 
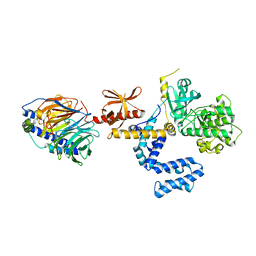 | | Bovine GRK2 in complex with Gbetagamma subunits | | Descriptor: | Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Thal, D.M, Tesmer, J.J. | | Deposit date: | 2010-12-01 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Molecular Mechanism of Selectivity among G Protein-Coupled Receptor Kinase 2 Inhibitors.
Mol.Pharmacol., 80, 2011
|
|
3PND
 
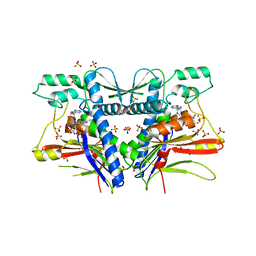 | | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD binding proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Thiamine biosynthesis lipoprotein ApbE | | Authors: | Boyd, J.M, Endrizzi, J.A, Hamilton, T.L, Christopherson, M.R, Mulder, D.W, Downs, D.M, Peters, J.W. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD-binding proteins.
J.Bacteriol., 193, 2011
|
|
3PVU
 
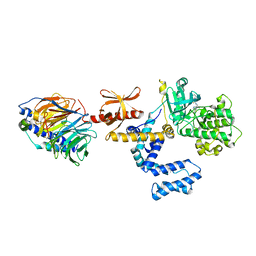 | | Bovine GRK2 in complex with Gbetagamma subunits and a selective kinase inhibitor (CMPD101) | | Descriptor: | 3-({[4-methyl-5-(pyridin-4-yl)-4H-1,2,4-triazol-3-yl]methyl}amino)-N-[2-(trifluoromethyl)benzyl]benzamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Thal, D.M, Tesmer, J.J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Molecular Mechanism of Selectivity among G Protein-Coupled Receptor Kinase 2 Inhibitors.
Mol.Pharmacol., 80, 2011
|
|
3QHR
 
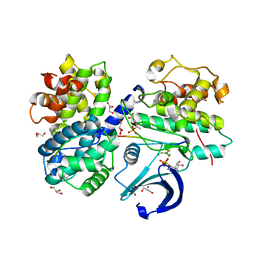 | | Structure of a pCDK2/CyclinA transition-state mimic | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CDK2 substrate peptide: PKTPKKAKKL, CHLORIDE ION, ... | | Authors: | Young, M.A, Jacobsen, D.M, Bao, Z.Q. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Briefly Bound to Activate: Transient Binding of a Second Catalytic Magnesium Activates the Structure and Dynamics of CDK2 Kinase for Catalysis.
Structure, 19, 2011
|
|
3O78
 
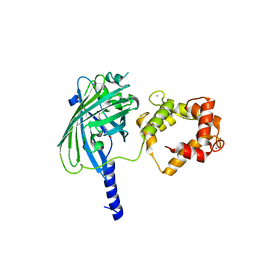 | | The structure of Ca2+ Sensor (Case-12) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, smooth muscle,Green fluorescent protein,Green fluorescent protein,Calmodulin-1 | | Authors: | Leder, L, Stark, W, Freuler, F, Marsh, M, Meyerhofer, M, Stettler, T, Mayr, L.M, Britanova, O.V, Strukova, L.A, Chudakov, D.M. | | Deposit date: | 2010-07-30 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of Ca2+ sensor Case16 reveals the mechanism of reaction to low Ca2+ concentrations
Sensors (Basel), 10, 2010
|
|
3PRV
 
 | | Nucleoside diphosphate kinase B from Trypanosoma cruzi | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Souza, T.A.C.B, Trindade, D.M, Tonoli, C.C.C, Santos, C.R, Oliveira, A.H.C, Murakami, M.T. | | Deposit date: | 2010-11-30 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Molecular adaptability of nucleoside diphosphate kinase b from trypanosomatid parasites: stability, oligomerization and structural determinants of nucleotide binding.
Mol Biosyst, 7, 2011
|
|
3PVW
 
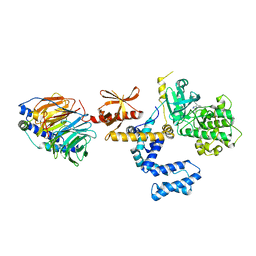 | |
