3MN2
 
 | |
2VV2
 
 | | hPPARgamma Ligand binding domain in complex with 5-HEPA | | Descriptor: | (5R,6E,8Z,11Z,14Z,17Z)-5-hydroxyicosa-6,8,11,14,17-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | Descriptor: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | Authors: | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
3OMK
 
 | |
3OKH
 
 | |
7NXZ
 
 | |
3NEU
 
 | |
2WUI
 
 | | Crystal Structure of MexZ, a key repressor responsible for antibiotic resistance in Pseudomonas aeruginosa. | | Descriptor: | TRANSCRIPTIONAL REGULATOR | | Authors: | Alguel, Y, Lu, D, Quade, N, Zhang, X. | | Deposit date: | 2009-10-05 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Mexz, a Key Repressor Responsible for Antibiotic Resistance in Pseudomonas Aeruginosa.
J.Struct.Biol., 172, 2010
|
|
2X4H
 
 | | Crystal Structure of the hypothetical protein SSo2273 from Sulfolobus solfataricus | | Descriptor: | HYPOTHETICAL PROTEIN SSO2273, ZINC ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-31 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
8EYT
 
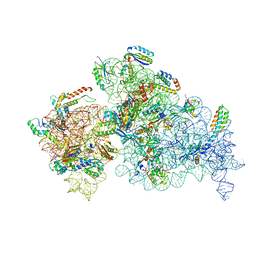 | | 30S_delta_ksgA+KsgA complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sun, J, Kinman, L.F, Jahagirdar, D, Ortega, J, Davis, J.H. | | Deposit date: | 2022-10-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | KsgA facilitates ribosomal small subunit maturation by proofreading a key structural lesion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2VSR
 
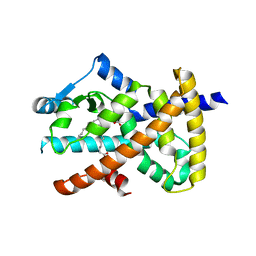 | | hPPARgamma Ligand binding domain in complex with 9-(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
3PAS
 
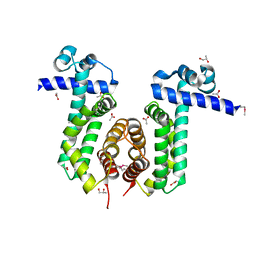 | |
4HV5
 
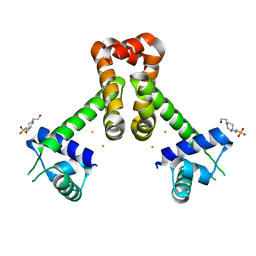 | | Structure of the MNTR FE2+ complex with E site metal binding | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FE (II) ION, Transcriptional regulator MntR | | Authors: | Glasfeld, A, Brophy, M.B, Kliegman, J.I, Griner, S.L. | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Roles of the A and C sites in the manganese-specific activation of MntR.
Biochemistry, 52, 2013
|
|
7NUF
 
 | | Vaccinia virus protein 018 in complex with STAT1 | | Descriptor: | ACETYL GROUP, SULFATE ION, Signal transducer and activator of transcription 1-alpha/beta, ... | | Authors: | Pantelejevs, T, Talbot-Cooper, C, Smith, G.L, Hyvonen, M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.0004015 Å) | | Cite: | Poxviruses and paramyxoviruses use a conserved mechanism of STAT1 antagonism to inhibit interferon signaling.
Cell Host Microbe, 30, 2022
|
|
4HX4
 
 | |
2OI4
 
 | |
4JAZ
 
 | | Crystal structure of the complex between PPARgamma LBD and trans-resveratrol | | Descriptor: | Peroxisome proliferator-activated receptor gamma, RESVERATROL | | Authors: | Pochetti, G, Capelli, D, Montanari, R, Calleri, E, Moaddel, R, Temporini, C. | | Deposit date: | 2013-02-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Resveratrol and Its Metabolites Bind to PPARs.
Chembiochem, 15, 2014
|
|
2VST
 
 | | hPPARgamma Ligand binding domain in complex with 13-(S)-HODE | | Descriptor: | (9Z,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV3
 
 | | hPPARgamma Ligand binding domain in complex with 4-oxoDHA | | Descriptor: | (6E,10Z,13Z,16Z,19Z)-4-oxodocosa-6,10,13,16,19-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation of Ppargamma by Oxidized Fatty Acids.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3QF9
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a furan-thiazolidinedione ligand | | Descriptor: | 6-{5-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]furan-2-yl}-N-{3-[(4-ethylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl}naphthalene-1-carboxamide, CHLORIDE ION, Proto-oncogene serine/threonine-protein kinase pim-1, ... | | Authors: | Filippakopoulos, P, Bullock, A.N, Fedorov, O, Miduturu, C.V, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Grey, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-21 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a furan-thiazolidinedione ligand
To be Published
|
|
2XYW
 
 | |
2V0V
 
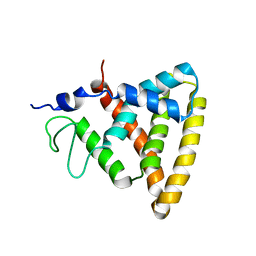 | | Crystal Structure of Rev-Erb beta | | Descriptor: | ORPHAN NUCLEAR RECEPTOR NR1D2 | | Authors: | Woo, E.-J, Jeong, D.G, Lim, M.-Y, Jun Kim, S, Eon Ryu, S. | | Deposit date: | 2007-05-19 | | Release date: | 2007-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into the Constitutive Repression Function of the Nuclear Receptor Rev-Erbbeta
J.Mol.Biol., 373, 2007
|
|
2XYX
 
 | |
3NQO
 
 | |
3R4K
 
 | |
