5I34
 
 | | Adenylosuccinate synthetase from Cryptococcus neoformans complexed with GDP and IMP | | Descriptor: | Adenylosuccinate synthetase, GUANOSINE-5'-DIPHOSPHATE, INOSINIC ACID | | Authors: | Blundell, R.D, Williams, S.J, Ericsson, D, Fraser, J.A, Kobe, B. | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Disruption of de Novo Adenosine Triphosphate (ATP) Biosynthesis Abolishes Virulence in Cryptococcus neoformans.
Acs Infect Dis., 2, 2016
|
|
3L61
 
 | | Crystal structure of substrate-free P450cam at 200 mM [K+] | | Descriptor: | Camphor 5-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, Y.-T, Wilson, R.F, Rupniewski, I, Goodin, D.B. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | P450cam visits an open conformation in the absence of substrate.
Biochemistry, 49, 2010
|
|
6RK9
 
 | |
2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-26 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
6P2H
 
 | |
7LLQ
 
 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | Descriptor: | 1-benzyl-4-methoxybenzene, 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, Leukotriene A-4 hydrolase, ... | | Authors: | Lee, K.H, Lee, S.H, Shim, Y.M, Paige, M, Noble, S.M. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
3L63
 
 | | Crystal structure of camphor-bound P450cam at low [K+] | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Rupniewski, I, Goodin, D.B. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | P450cam visits an open conformation in the absence of substrate.
Biochemistry, 49, 2010
|
|
3P8T
 
 | | Crystal structure of the archaeal asparagine synthetase A | | Descriptor: | AsnS-like asparaginyl-tRNA synthetase related protein | | Authors: | Blaise, M, Kern, D. | | Deposit date: | 2010-10-15 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Archaeal Asparagine Synthetase: Interrelation with Aspartyl-tRNA and Asparaginyl-tRNA Synthetases.
J.Mol.Biol., 412, 2011
|
|
1I2T
 
 | |
8JA8
 
 | |
4BVY
 
 | |
5LVS
 
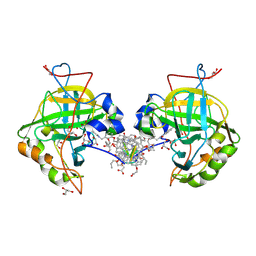 | | Self-assembled protein-aromatic foldamer complexes with 2:3 and 2:2:1 stoichiometries | | Descriptor: | Aromatic foldamer, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Jewginski, M, LANGLOIS D'ESTAINTOT, B, Granier, T, Huc, Y. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
7OJT
 
 | | Crystal structure of unliganded PatA, a membrane associated acyltransferase from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Phosphatidylinositol mannoside acyltransferase | | Authors: | Anso, I, Wang, L, Marina, A, Paez-Perez, E.D, Perrone, S, Lowary, T.L, Trastoy, B, Guerin, M.E. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.67 Å) | | Cite: | Molecular ruler mechanism and interfacial catalysis of the integral membrane acyltransferase PatA.
Sci Adv, 7, 2021
|
|
7P76
 
 | | Re-engineered 2-deoxy-D-ribose-5-phosphate aldolase catalysing asymmetric Michael addition reactions, Schiff base complex with cinnamaldehyde | | Descriptor: | (2E)-3-phenylprop-2-enal, Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Thunnissen, A.M.W.H, Rozeboom, H.J, Kunzendorf, A, Poelarends, G.J. | | Deposit date: | 2021-07-19 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unlocking Asymmetric Michael Additions in an Archetypical Class I Aldolase by Directed Evolution.
Acs Catalysis, 11, 2021
|
|
6RKT
 
 | | Crystal Structure of TGT in complex with N2-methyl-1H,7H,8H-imidazo[4,5-g]quinazoline-2,6-diamine | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Hassaan, E, Heine, A, Klebe, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Fragment Screening Hit Draws Attention to a Novel Transient Pocket Adjacent to the Recognition Site of the tRNA-Modifying Enzyme TGT.
J.Med.Chem., 63, 2020
|
|
3U8K
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3573 (1-(5-ethoxypyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
6S2Y
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-soluble chlorophyll protein | | Authors: | Agostini, A, Meneghin, E, Gewehr, L, Pedron, D, Palm, D.M, Carbonera, D, Paulsen, H, Jaenicke, E, Collini, E. | | Deposit date: | 2019-06-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How water-mediated hydrogen bonds affect chlorophyll a/b selectivity in Water-Soluble Chlorophyll Protein.
Sci Rep, 9, 2019
|
|
7P75
 
 | |
3LA2
 
 | | Crystal structure of NtcA in complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Global nitrogen regulator | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Chen, Y.X, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7OSA
 
 | | Pre-translocation complex of 80 S.cerevisiae ribosome with eEF2 and ligands | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Djumagulov, M, Jenner, L, Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2021-06-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accuracy mechanism of eukaryotic ribosome translocation.
Nature, 600, 2021
|
|
3P8V
 
 | |
7OSM
 
 | | Intermediate translocation complex of 80 S.cerevisiae ribosome with eEF2 and ligands | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Djumagulov, M, Jenner, L, Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accuracy mechanism of eukaryotic ribosome translocation.
Nature, 600, 2021
|
|
3U8N
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3950 (1-(6-bromo-5-ethoxypyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(6-bromo-5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
2ROC
 
 | | Solution structure of Mcl-1 Complexed with Puma | | Descriptor: | Bcl-2-binding component 3, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
2ROD
 
 | | Solution Structure of MCL-1 Complexed with NoxaA | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Noxa | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
