8ASV
 
 | | Cryo-EM structure of yeast Elongator complex | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
3FF2
 
 | |
6OVT
 
 | | Crystal Structure of IlvD from Mycobacterium tuberculosis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dihydroxy-acid dehydratase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Almo, S.C, Grove, T.L, Bonanno, J.B, Baker, E.N, Bashiri, G. | | Deposit date: | 2019-05-08 | | Release date: | 2019-08-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The active site of theMycobacterium tuberculosisbranched-chain amino acid biosynthesis enzyme dihydroxyacid dehydratase contains a 2Fe-2S cluster.
J.Biol.Chem., 294, 2019
|
|
6OWG
 
 | | Structure of a synthetic beta-carboxysome shell, T=4 | | Descriptor: | Ethanolamine utilization protein EutN/carboxysome structural protein Ccml, Microcompartments protein | | Authors: | Sutter, M, Laughlin, T.G, Davies, K.M, Kerfeld, C.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a Syntheticbeta-Carboxysome Shell.
Plant Physiol., 181, 2019
|
|
7C6C
 
 | | Crystal structure of native chitosanase from Bacillus subtilis MY002 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Chitosanase | | Authors: | Gou, Y, Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.258 Å) | | Cite: | Structure-based rational design of chitosanase CsnMY002 for high yields of chitobiose.
Colloids Surf B Biointerfaces, 202, 2021
|
|
7C6D
 
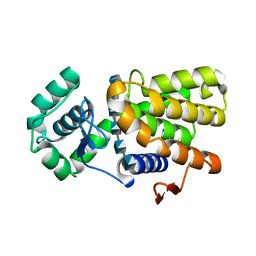 | | Crystal structure of E19A mutant chitosanase from Bacillus subtilis MY002 complexed with 6 GlcN. | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Chitosanase | | Authors: | Gou, Y, Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structure-based rational design of chitosanase CsnMY002 for high yields of chitobiose.
Colloids Surf B Biointerfaces, 202, 2021
|
|
6P7L
 
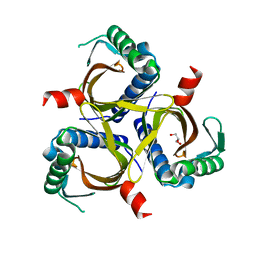 | |
6IFK
 
 | | Cryo-EM structure of type III-A Csm-CTR1 complex, AMPPNP bound | | Descriptor: | CTR1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6XQE
 
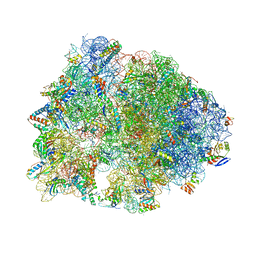 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with sarecycline, UAA-mRNA, and deacylated P-site tRNA at 3.00A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Lomakin, I.B, Bunick, C.G, Polikanov, Y.S. | | Deposit date: | 2020-07-09 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Sarecycline interferes with tRNA accommodation and tethers mRNA to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XQD
 
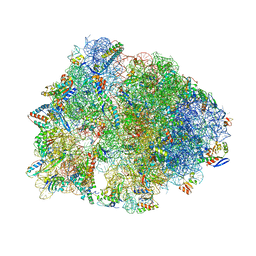 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with sarecycline, UUC-mRNA, and deacylated P-site tRNA at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Lomakin, I.B, Bunick, C.G, Polikanov, Y.S. | | Deposit date: | 2020-07-09 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sarecycline interferes with tRNA accommodation and tethers mRNA to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5EUM
 
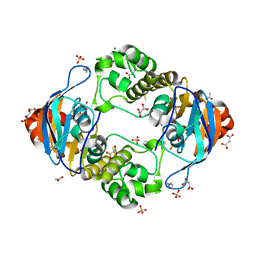 | | 1.8 Angstrom Crystal Structure of ATP-binding Component of Fused Lipid Transporter Subunits of ABC superfamily from Haemophilus influenzae. | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipid A export ATP-binding/permease protein MsbA, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of ATP-binding Component of Fused Lipid Transporter Subunits of ABC superfamily from Haemophilus influenzae.
To Be Published
|
|
6IG0
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1, ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTR1, MAGNESIUM ION, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
8G64
 
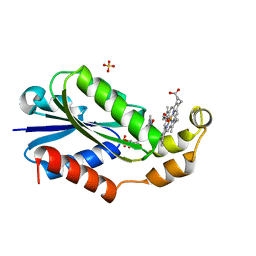 | | Heme-bound flavodoxin FldH from Fusobacterium nucleatum | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chan, A.C, Wolthers, K.R, Murphy, M.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new member of the flavodoxin superfamily from Fusobacterium nucleatum that functions in heme trafficking and reduction of anaerobilin.
J.Biol.Chem., 299, 2023
|
|
6XI9
 
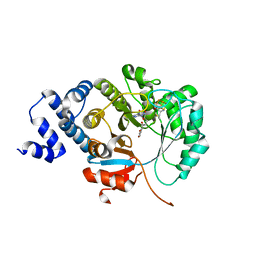 | | X-ray crystal structure of MqnE from Pedobacter heparinus in complex with aminofutalosine and methionine | | Descriptor: | 9-[7-(3-carboxyphenyl)-5,6-dideoxy-beta-D-ribo-heptodialdo-1,4-furanosyl]-9H-purin-6-amine, Aminodeoxyfutalosine synthase, CHLORIDE ION, ... | | Authors: | Grove, T.L, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2020-06-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Narrow-Spectrum Antibiotic Targeting of the Radical SAM Enzyme MqnE in Menaquinone Biosynthesis.
Biochemistry, 59, 2020
|
|
7MRU
 
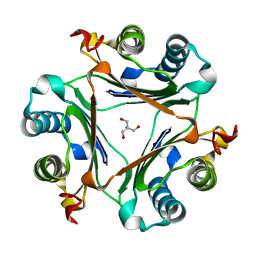 | | Crystal structure of S62A MIF2 mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-dopachrome decarboxylase | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MSE
 
 | | High-resolution crystal structure of hMIF2 with tartrate at the active site | | Descriptor: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MRV
 
 | | F100A mutant structure of MIF2 (D-DT) | | Descriptor: | D-dopachrome decarboxylase, SULFATE ION | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
6PQ1
 
 | | Structure of the Fremyella diplosiphon OCP1 | | Descriptor: | Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | Authors: | Sutter, M, Dominguez-Martin, M.A, Bao, H, Kerfeld, C.A. | | Deposit date: | 2019-07-08 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Comparative ultrafast spectroscopy and structural analysis of OCP1 and OCP2 from Tolypothrix.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
5EL1
 
 | |
6NLU
 
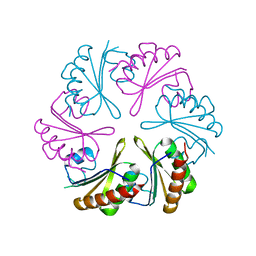 | |
7M5O
 
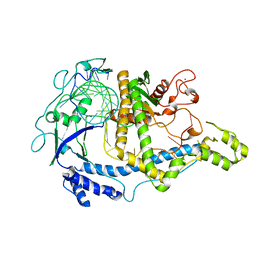 | | Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA | | Descriptor: | CasPhi, ZINC ION, crRNA | | Authors: | Pausch, P, Soczek, K, Nogales, E, Doudna, J. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | DNA interference states of the hypercompact CRISPR-Cas Phi effector.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6Y6L
 
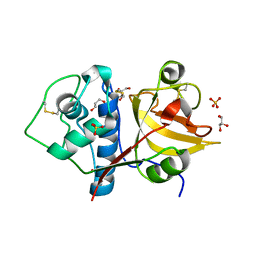 | | Structure the ananain protease from Ananas comosus with a thiomethylated catalytic cysteine | | Descriptor: | Ananain, GLYCEROL, SULFATE ION | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6YCB
 
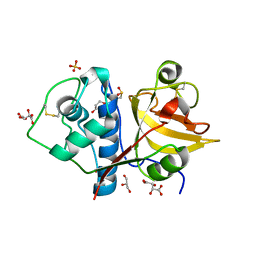 | | Structure the ananain protease from Ananas comosus covalently bound to with the E64 inhibitor | | Descriptor: | Ananain, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.257 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
7LYS
 
 | | Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA and DNA | | Descriptor: | CasPhi-2, NTS-DNA, TS-DNA, ... | | Authors: | Pausch, P, Soczek, K, Nogales, E, Doudna, J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | DNA interference states of the hypercompact CRISPR-Cas Phi effector.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LYT
 
 | | Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA and Phosphorothioate-DNA | | Descriptor: | CasPhi, MAGNESIUM ION, NTS-DNA*, ... | | Authors: | Pausch, P, Soczek, K, Nogales, E, Doudna, J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | DNA interference states of the hypercompact CRISPR-Cas Phi effector.
Nat.Struct.Mol.Biol., 28, 2021
|
|
