6MPV
 
 | |
7TL0
 
 | | Cryo-EM structure of hMPV preF bound by Fabs MPE8 and SAN32-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Rush, S.A, Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-09-14 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Characterization of prefusion-F-specific antibodies elicited by natural infection with human metapneumovirus.
Cell Rep, 40, 2022
|
|
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | Descriptor: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | Deposit date: | 2018-12-15 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
8GFV
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #1 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
6NR4
 
 | | Cryo-EM structure of the TRPM8 ion channel with low occupancy icilin, PI(4,5)P2, and calcium | | Descriptor: | Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
1I5H
 
 | |
4B2B
 
 | | Structure of the factor Xa-like trypsin variant triple-Ala (TGPA) in complex with eglin C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Menzel, A, Neumann, P, Stubbs, M.T. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Thermodynamic signatures in macromolecular interactions involving conformational flexibility.
Biol.Chem., 395, 2014
|
|
6NBC
 
 | |
6NR2
 
 | | Cryo-EM structure of the TRPM8 ion channel in complex with the menthol analog WS-12 and PI(4,5)P2 | | Descriptor: | (1R,2S,5R)-N-(4-methoxyphenyl)-5-methyl-2-(propan-2-yl)cyclohexane-1-carboxamide, (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
1XY8
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
6N09
 
 | |
5IFM
 
 | | Human NONO (p54nrb) Homodimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-POU domain-containing octamer-binding protein, ... | | Authors: | Knott, G.J, Bond, C.S. | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic study of human NONO (p54(nrb)): overcoming pathological problems with purification, data collection and noncrystallographic symmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8GBD
 
 | | Homo sapiens Zalpha mutant - P193A | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Langeberg, C.J, Vogeli, B, Nichols, P.J, Henen, M, Vicens, Q. | | Deposit date: | 2023-02-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential Structural Features of Two Mutant ADAR1p150 Z alpha Domains Associated with Aicardi-Goutieres Syndrome.
J.Mol.Biol., 435, 2023
|
|
8POP
 
 | | HK97 small terminase in complex with DNA | | Descriptor: | DNA (31-MER), Terminase small subunit | | Authors: | Chechik, M, Greive, S.J, Antson, A.A, Jenkins, H.T. | | Deposit date: | 2023-07-05 | | Release date: | 2023-08-09 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for DNA recognition by a viral genome-packaging machine.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8GBC
 
 | | Homo sapiens Zalpha mutant - N173S | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Langeberg, C.J, Nichols, P.J, Henen, M, Vicens, Q, Vogeli, B. | | Deposit date: | 2023-02-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential Structural Features of Two Mutant ADAR1p150 Z alpha Domains Associated with Aicardi-Goutieres Syndrome.
J.Mol.Biol., 435, 2023
|
|
8G0Y
 
 | |
6NE3
 
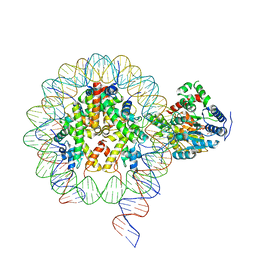 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h bound at SHL-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (156-MER), Histone H2A type 1, ... | | Authors: | Armache, J.-P, Gamarra, N, Johnson, S.L, Leonard, J.D, Wu, S, Narlikar, G.N, Cheng, Y. | | Deposit date: | 2018-12-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome.
Elife, 8, 2019
|
|
8G0X
 
 | |
5IRZ
 
 | | Structure of TRPV1 determined in lipid nanodisc | | Descriptor: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4S,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(pentanoyloxy)propan-2-yl decanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, (4R,7S)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(pentanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphatetradecan-1-aminium, ... | | Authors: | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | Deposit date: | 2016-03-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
1XXZ
 
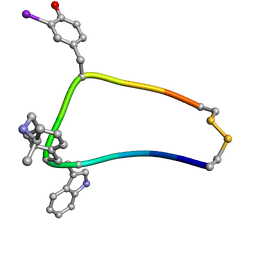 | | Solution structure of sst1-selective somatostatin (SRIF) analog | | Descriptor: | SST1-selective somatosatin (analog 5) | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
1XY4
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
3CQV
 
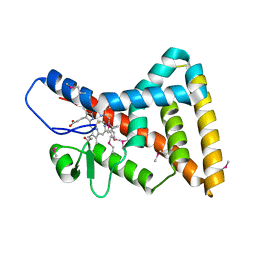 | | Crystal structure of Reverb beta in complex with heme | | Descriptor: | Nuclear receptor subfamily 1 group D member 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, X, Dong, A, Pardee, K.I, Reinking, J, Krause, H, Schuetz, A, Zhang, R, Cui, H, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Savchenko, A, Botchkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-03 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta.
Plos Biol., 7, 2009
|
|
6TFK
 
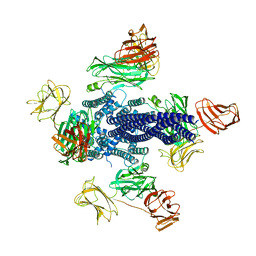 | | Vip3Aa toxin structure | | Descriptor: | MAGNESIUM ION, Vegetative insecticidal protein | | Authors: | Nunez-Ramirez, R, Huesa, J, Bel, Y, Ferre, J, Casino, P, Arias-Palomo, E. | | Deposit date: | 2019-11-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular architecture and activation of the insecticidal protein Vip3Aa from Bacillus thuringiensis.
Nat Commun, 11, 2020
|
|
1MV3
 
 | |
1N20
 
 | | (+)-Bornyl Diphosphate Synthase: Complex with Mg and 3-aza-2,3-dihydrogeranyl diphosphate | | Descriptor: | (+)-bornyl diphosphate synthase, 2-[METHYL-(4-METHYL-PENT-3-ENYL)-AMINO]-ETHYL-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bornyl Diphosphate Synthase: Structure and Strategy for Carbocation Manipulation by a Terpenoid Cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
