8AI2
 
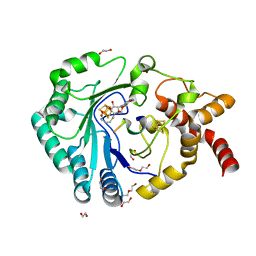 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
5NOV
 
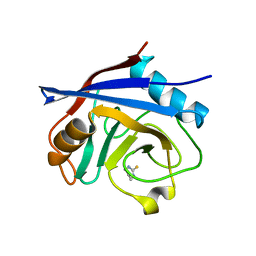 | | Structure of cyclophilin A in complex with hexahydropyrimidine-2-thione | | Descriptor: | 1,3-diazinane-2-thione, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
4WUA
 
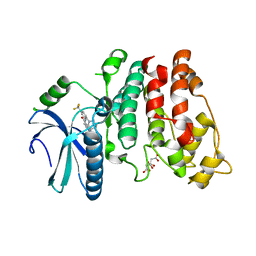 | | Crystal structure of human SRPK1 complexed to an inhibitor SRPIN340 | | Descriptor: | CITRIC ACID, N-[2-(1-piperidinyl)-5-(trifluoromethyl)phenyl]-4-pyridinecarboxamide, SRSF protein kinase 1, ... | | Authors: | Hoshina, M, Ikura, T, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Dual Inhibitor of SRPK1 and CK2 That Attenuates Pathological Angiogenesis of Macular Degeneration in Mice
Mol.Pharmacol., 88, 2015
|
|
6QIU
 
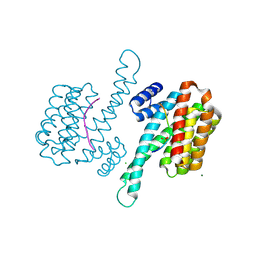 | | Crystal structure of 14-3-3 sigma in complex with Ataxin-1 Ser776 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Ataxin-1 phosphopeptide, CHLORIDE ION, ... | | Authors: | Leysen, S, Milroy, L.G, Davis, J.M, Brunsveld, L, Ottmann, C. | | Deposit date: | 2019-01-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural insights into the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1
To Be Published
|
|
4X7K
 
 | | Co-crystal Structure of PERK bound to 4-{2-amino-3-[5-fluoro-2-(methylamino)quinazolin-6-yl]-4-methylbenzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one inhibitor | | Descriptor: | 4-{2-amino-3-[5-fluoro-2-(methylamino)quinazolin-6-yl]-4-methylbenzoyl}-1-methyl-2,5-diphenyl-1,2-dihydro-3H-pyrazol-3-one, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL, ... | | Authors: | Shaffer, P.L, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
9L3S
 
 | | Staphylococcus aureus lipase-Penfluridol complex (in space) | | Descriptor: | 1-[4,4-bis(4-fluorophenyl)butyl]-4-[4-chloranyl-3-(trifluoromethyl)phenyl]piperidin-4-ol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Hirokawa, T, Kamo, M, Furubayasi, N, Inaka, K, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2024-12-19 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural analysis shows the mode of inhibition for Staphylococcus aureus lipase by antipsychotic penfluridol.
Sci Rep, 15, 2025
|
|
6QFC
 
 | | Structure of an anti-Mcl1 scFv | | Descriptor: | DIMETHYL SULFOXIDE, Induced myeloid leukemia cell differentiation protein Mcl-1, scFv55 | | Authors: | Luptak, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QGP
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-0769 | | Descriptor: | 1-cycloheptyl-3-[3-(cyclopentyloxy)-4-methoxyphenyl]-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-0769
To be published
|
|
4X0Y
 
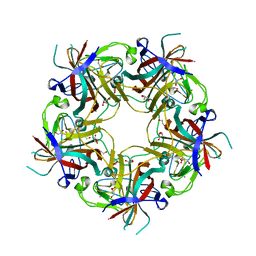 | | JC polyomavirus VP1 from a genotype 3 strain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein, ... | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Greater Affinity of JC Polyomavirus Capsid for alpha 2,6-Linked Lactoseries Tetrasaccharide c than for Other Sialylated Glycans Is a Major Determinant of Infectivity.
J.Virol., 89, 2015
|
|
7RBQ
 
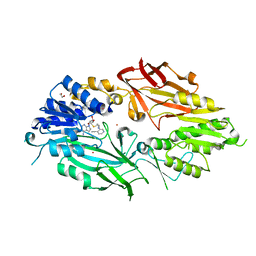 | | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Protein arginine N-methyltransferase 9, ... | | Authors: | Zeng, H, Dong, A, Hutchinson, A, Seitova, A, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Co-crystal structure of human PRMT9 in complex with MT556 inhibitor
To Be Published
|
|
7P7T
 
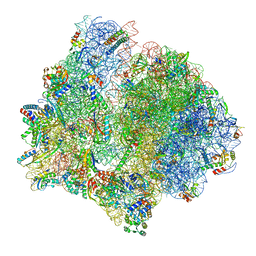 | | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state III | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7B0X
 
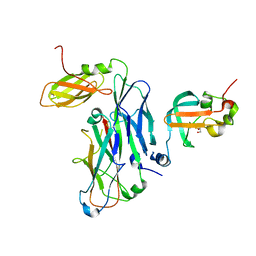 | | Crystal structure of the ternary complex of the E. coli type 1 pilus proteins FimC, FimI and the N-terminal domain of FimD | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, Fimbrin-like protein FimI, ... | | Authors: | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
7P7Q
 
 | | E. faecalis 70S ribosome bound by PoxtA-EQ2, high-resolution combined volume | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7P7S
 
 | | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state II | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
9CPJ
 
 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Method: | SOLUTION NMR | | Cite: | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1.
Bioorg.Med.Chem.Lett., 2024
|
|
9CPG
 
 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-carbamimidamidophenyl 4-carbamimidamidobenzoate, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Method: | SOLUTION NMR | | Cite: | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1.
Bioorg.Med.Chem.Lett., 2024
|
|
6QL1
 
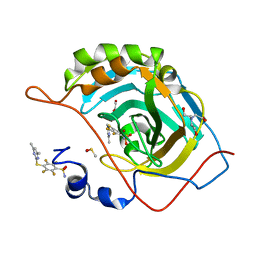 | | Crystal structure of chimeric carbonic anhydrase VI with 4-[(4,6-dimethylpyrimidin-2-yl)thio]-2,3,5,6-tetrafluorobenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4,6-dimethylpyrimidin-2-yl)thio]-2,3,5,6-tetrafluorobenzenesulfonamide, BICINE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-01-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Engineered Carbonic Anhydrase VI-Mimic Enzyme Switched the Structure and Affinities of Inhibitors.
Sci Rep, 9, 2019
|
|
9L3C
 
 | | Staphylococcus aureus lipase-Penfluridol complex (on the ground) | | Descriptor: | 1-[4,4-bis(4-fluorophenyl)butyl]-4-[4-chloranyl-3-(trifluoromethyl)phenyl]piperidin-4-ol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Hirokawa, T, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2024-12-18 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural analysis shows the mode of inhibition for Staphylococcus aureus lipase by antipsychotic penfluridol.
Sci Rep, 15, 2025
|
|
8DF5
 
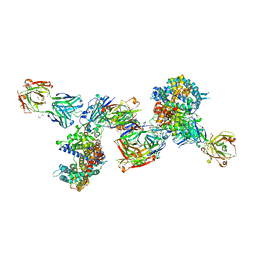 | | SARS-CoV-2 Beta RBD in complex with human ACE2 and S304 Fab and S309 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | McCallum, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution.
Science, 377, 2022
|
|
8FBT
 
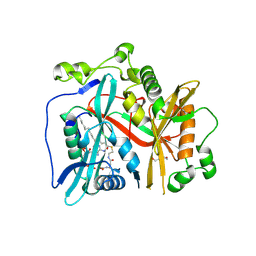 | | Crystal structure of Cryptosporidium parvum N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Staker, B.L, Fenwick, M.K, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of and Structural Insights into Hit Compounds Targeting N -Myristoyltransferase for Cryptosporidium Drug Development.
Acs Infect Dis., 9, 2023
|
|
6WXC
 
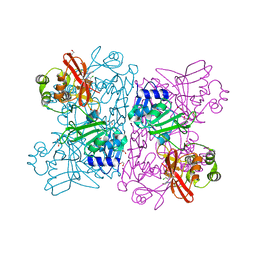 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with potential repurposing drug Tipiracil | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
8Z69
 
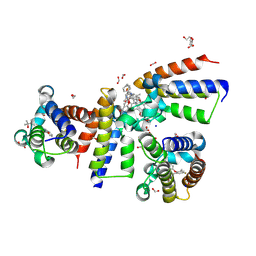 | | Crystal Structure of the second bromodomain of human BRD2 BD2 in complex with the inhibitor Y13195 | | Descriptor: | 1,2-ETHANEDIOL, 7-(2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl)-5-methyl-2-(2-phenyl-1H-imidazol-5-yl)furo[3,2-c]pyridin-4(5H)-one, BRD2_HUMAN, ... | | Authors: | Li, J, Hu, Q, Xu, H, Zhao, X, Zhang, C, Zhu, R, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2024-04-18 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of the First BRD4 Second Bromodomain (BD2)-Selective Inhibitors.
J.Med.Chem., 67, 2024
|
|
8F5U
 
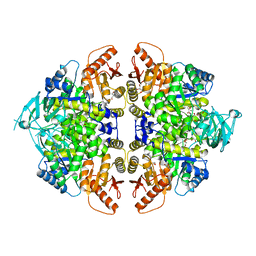 | | Rabbit muscle pyruvate kinase in complex with magnesium, potassium and pyruvate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
5XII
 
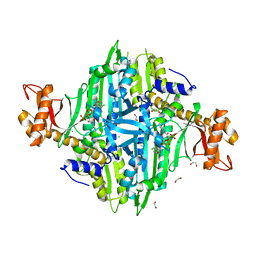 | | Crystal Structure of Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in complex with inhibitor 6 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranyl-7-fluoranyl-3-[3-[(2R)-3-oxidanylidenepiperidin-2-yl]propyl]quinazolin-4-one, CHLORIDE ION, ... | | Authors: | Jain, V, Manickam, Y, Sharma, A. | | Deposit date: | 2017-04-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Targeting Prolyl-tRNA Synthetase to Accelerate Drug Discovery against Malaria, Leishmaniasis, Toxoplasmosis, Cryptosporidiosis, and Coccidiosis
Structure, 25, 2017
|
|
8ESM
 
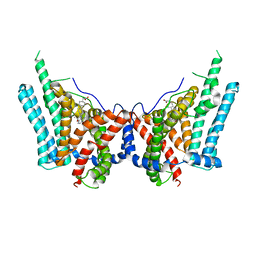 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with T863 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, {(1r,4r)-4-[4-(4-amino-7,7-dimethyl-7H-pyrimido[4,5-b][1,4]oxazin-6-yl)phenyl]cyclohexyl}acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
