6CHP
 
 | |
2AU9
 
 | | Inorganic pyrophosphatase complexed with substrate | | Descriptor: | CHLORIDE ION, FLUORIDE ION, Inorganic pyrophosphatase, ... | | Authors: | Samygina, V.R, Popov, A.N, Avaeva, S.M, Bartunik, H.D. | | Deposit date: | 2005-08-27 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies
J.Mol.Biol., 366, 2007
|
|
4J22
 
 | | Tankyrase 2 in complex with 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide | | Descriptor: | 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
5N6W
 
 | | Retinoschisin R141H Mutant | | Descriptor: | Retinoschisin | | Authors: | Ramsay, E.P, Collins, R.F, Owens, T.W, Siebert, C.A, Jones, R.P.O, Roseman, A, Wang, T, Baldock, C. | | Deposit date: | 2017-02-16 | | Release date: | 2017-04-12 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of X-linked retinoschisis mutations reveals distinct classes which differentially effect retinoschisin function
Human Molecular Genetics, 25, 2016
|
|
2AKH
 
 | | Normal mode-based flexible fitted coordinates of a non-translocating SecYEG protein-conducting channel into the cryo-EM map of a SecYEG-nascent chain-70S ribosome complex from E. coli | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, Protein-export membrane protein secG | | Authors: | Mitra, K.M, Schaffitzel, C, Shaikh, T, Tama, F, Jenni, S, Brooks III, C.L, Ban, N, Frank, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | Structure of the E. coli protein-conducting channel bound to a translating ribosome.
Nature, 438, 2005
|
|
6BVB
 
 | | Crystal structure of HIF-2alpha-pVHL-elongin B-elongin C | | Descriptor: | Elongin-B, Elongin-C, Hypoxia-Inducible Factor 2 alpha, ... | | Authors: | Tarade, D, Ohh, M, Lee, J.E. | | Deposit date: | 2017-12-12 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | HIF-2 alpha-pVHL complex reveals broad genotype-phenotype correlations in HIF-2 alpha-driven disease.
Nat Commun, 9, 2018
|
|
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
2JKS
 
 | | Crystal structure of the the bradyzoite specific antigen BSR4 from toxoplasma gondii. | | Descriptor: | BRADYZOITE SURFACE ANTIGEN BSR4, ZINC ION | | Authors: | Boulanger, M.J, Grigg, M.E, Bruic, E, Grujic, O. | | Deposit date: | 2008-08-29 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of the Bradyzoite Surface Antigen (Bsr4) from Toxoplasma Gondii, a Unique Addition to the Surface Antigen Glycoprotein 1-Related Superfamily.
J.Biol.Chem., 284, 2009
|
|
4IUE
 
 | | Tankyrase in complex with 7-(2-fluorophenyl)-4-methyl-1,2-dihydroquinolin-2-one | | Descriptor: | 7-(2-fluorophenyl)-4-methylquinolin-2(1H)-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-01-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4A6V
 
 | | X-ray structures of oxazole hydroxamate EcMetAp-Mn complexes | | Descriptor: | CARBONATE ION, MANGANESE (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Huguet, F, Melet, A, AlvesdeSousa, R, Lieutaud, A, Chevalier, J, Deschamps, P, Tomas, A, Leulliot, N, Pages, J.M, Artaud, I. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Hydroxamic Acids as Potent Inhibitors of Fe(II) and Mn(II) E. Coli Methionine Aminopeptidase: Biological Activities and X-Ray Structures of Oxazole Hydroxamate-Ecmetap-Mn Complexes.
Chemmedchem, 7, 2012
|
|
1O1W
 
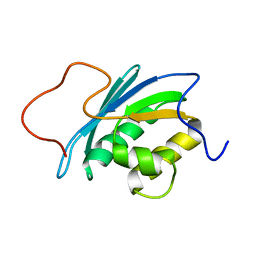 | | SOLUTION STRUCTURE OF THE RNASE H DOMAIN OF THE HIV-1 REVERSE TRANSCRIPTASE IN THE PRESENCE OF MAGNESIUM | | Descriptor: | RIBONUCLEASE H | | Authors: | Pari, K, Mueller, G.A, Derose, E.F, Kirby, T.W, London, R.E. | | Deposit date: | 2003-02-12 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Rnase H Domain of the HIV-1 Reverse Transcriptase in the Presence of Magnesium
Biochemistry, 42, 2003
|
|
2ZC2
 
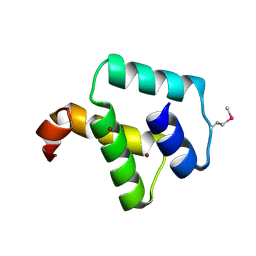 | | Crystal structure of DnaD-like replication protein from Streptococcus mutans UA159, gi 24377835, residues 127-199 | | Descriptor: | DnaD-like replication protein, ZINC ION | | Authors: | Duke, N.E.C, Clancy, S, Duggan, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of DnaD-like replication protein from Streptococcus mutans UA159.
To be Published
|
|
1TGE
 
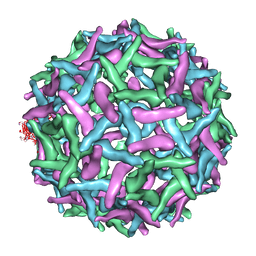 | | The structure of immature Dengue virus at 12.5 angstrom | | Descriptor: | envelope glycoprotein | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-05-28 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein.
Structure, 12, 2004
|
|
5GR3
 
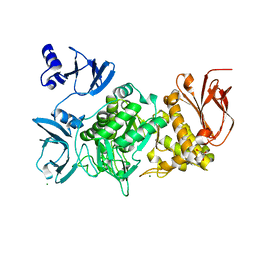 | |
6BYB
 
 | | Crystal structure of L3MBTL1 MBT Domain with MBK14970 | | Descriptor: | (S)-N-(cyclopropylmethyl)-N~2~-methyl-N-[2-methyl-2-(1-methylpiperidin-4-yl)propyl]alaninamide, 1,2-ETHANEDIOL, Lethal(3)malignant brain tumor-like protein 1, ... | | Authors: | DONG, A, DOBROVETSKY, E, NICHOLSON, B, COX, C, FISCHER, C, ARMACOST, K, SANDERS, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of L3MBTL1 MBT Domain with MBK14970
to be published
|
|
1ZWJ
 
 | | X-ray structure of galt-like protein from arabidopsis thaliana AT5G18200 | | Descriptor: | ZINC ION, putative galactose-1-phosphate uridyl transferase | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, McCoy, J.G, Johnson, K.A, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of an ADP-Glucose Phosphorylase from
Arabidopsis thaliana
Biochemistry, 45, 2006
|
|
4AF0
 
 | | Crystal structure of cryptococcal inosine monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, MYCOPHENOLIC ACID, ... | | Authors: | Valkov, E, Stamp, A, Morrow, C.A, Kobe, B, Fraser, J.A. | | Deposit date: | 2012-01-15 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De Novo GTP Biosynthesis is Critical for Virulence of the Fungal Pathogen Cryptococcus Neoformans
Plos Pathog., 8, 2012
|
|
7L11
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 2-[3-(3-chloro-5-propoxyphenyl)-2-oxo[2H-[1,3'-bipyridine]]-5-yl]benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
1ZYJ
 
 | | Human P38 MAP Kinase in Complex with Inhibitor 1a | | Descriptor: | 4-PHENOXY-N-(PYRIDIN-2-YLMETHYL)BENZAMIDE, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Karpusas, M, Michelotti, E.L, Springman, E.B. | | Deposit date: | 2005-06-10 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two classes of p38alpha MAP kinase inhibitors having a common diphenylether core but exhibiting divergent binding modes.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5UWC
 
 | | Cytokine-receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Broughton, S.E, Parker, M.W. | | Deposit date: | 2017-02-21 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dual role for the N-terminal domain of the IL-3 receptor in cell signalling.
Nat Commun, 9, 2018
|
|
7XZ5
 
 | | GPR119-Gs-LPC complex | | Descriptor: | (4R,7R,18E)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1ZZ2
 
 | | Two Classes of p38alpha MAP Kinase Inhibitors Having a Common Diphenylether Core but Exhibiting Divergent Binding Modes | | Descriptor: | Mitogen-activated protein kinase 14, N-[3-(4-FLUOROPHENOXY)PHENYL]-4-[(2-HYDROXYBENZYL)AMINO]PIPERIDINE-1-SULFONAMIDE, octyl beta-D-glucopyranoside | | Authors: | Michelotti, E.L, Moffett, K.K, Springman, E.B, Karpusas, M. | | Deposit date: | 2005-06-13 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two classes of p38alpha MAP kinase inhibitors having a common diphenylether core but exhibiting divergent binding modes.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
7KGF
 
 | |
7KVE
 
 | |
7KXY
 
 | |
