6ZF2
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[(4-propylphenyl)sulfonylamino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF3
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, SULFATE ION, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF7
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-hydroxy-2-oxoethyl-(3-methoxyphenyl)sulfonyl-amino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF0
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[(2,3,5,6-tetramethylphenyl)sulfonylamino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
4TQH
 
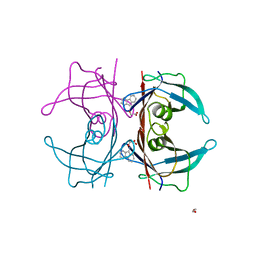 | | Human transthyretin (TTR) complexed with 3-(9H-fluoren-9-ylideneaminooxy)ethanoic acid | | Descriptor: | 1,2-ETHANEDIOL, Transthyretin, [(9H-fluoren-9-ylideneamino)oxy]acetic acid | | Authors: | Ciccone, L, Nencetti, S, Rossello, A, Orlandini, E, Stura, E.A. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | X-ray crystal structure and activity of fluorenyl-based compounds as transthyretin fibrillogenesis inhibitors.
J Enzyme Inhib Med Chem, 2015
|
|
6L69
 
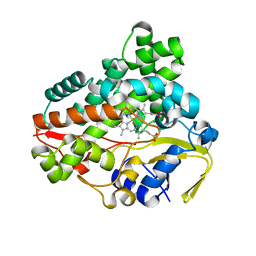 | | Crystal structure of CYP154C2 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regio- and stereoselective hydroxylation of testosterone by a novel cytochrome P450 154C2 from Streptomyces avermitilis.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
8BIN
 
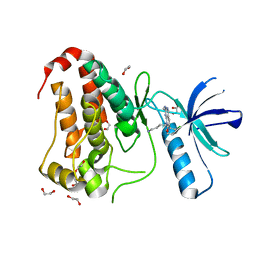 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with MR21 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-(2-chlorophenyl)-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Ephrin type-A receptor 2 | | Authors: | Zhubi, R, Rak, M, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Back-Pocket Optimization of 2-Aminopyrimidine-Based Macrocycles Leads to Potent EPHA2/GAK Kinase Inhibitors.
J.Med.Chem., 2024
|
|
4EK1
 
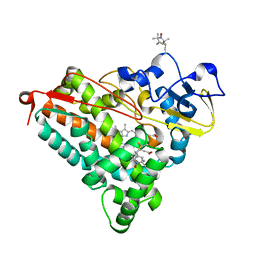 | | Crystal Structure of Electron-Spin Labeled Cytochrome P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Goodin, D.B. | | Deposit date: | 2012-04-08 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Double electron-electron resonance shows cytochrome P450cam undergoes a conformational change in solution upon binding substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EX8
 
 | | Crystal structure of the prealnumycin C-glycosynthase AlnA | | Descriptor: | AlnA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6Z9X
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting LLS (t-butyl)Y FGTPT | | Descriptor: | Beta-2-microglobulin, LEU-LEU-SER-TUR-PHE-GLY-THR-PRO-THR, MHC class I antigen, ... | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
4E83
 
 | |
4EIY
 
 | | Crystal structure of the chimeric protein of A2aAR-BRIL in complex with ZM241385 at 1.8A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Liu, W, Chun, E, Thompson, A.A, Chubukov, P, Xu, F, Katritch, V, Han, G.W, Heitman, L.H, Ijzerman, A.P, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-04-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for allosteric regulation of GPCRs by sodium ions.
Science, 337, 2012
|
|
4E82
 
 | |
8BLT
 
 | | Structure of Lactobacillus salivarius (Ls) bile salt hydrolase(BSH) in complex with taurocholate (TCA) | | Descriptor: | Bile salt hydrolase, TAUROCHOLIC ACID | | Authors: | Karlov, D.S, Long, S.L, Zeng, X, Xu, F, Lal, K, Cao, L, Hayoun, K, Lin, J, Joyce, S.A, Tikhonova, I.G. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the mechanism of bile salt hydrolase substrate specificity by experimental and computational analyses.
Structure, 31, 2023
|
|
8BLS
 
 | | Structure of Lactobacillus salivarius (Ls) bile salt hydrolase(BSH) in complex with Glycocholate (GCA) | | Descriptor: | Bile salt hydrolase, GLYCOCHOLIC ACID | | Authors: | Karlov, D.S, Long, S.L, Zeng, X, Xu, F, Lal, K, Cao, L, Hayoun, K, Lin, J, Joyce, S.A, Tikhonova, I.G. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the mechanism of bile salt hydrolase substrate specificity by experimental and computational analyses.
Structure, 31, 2023
|
|
4WCI
 
 | | Crystal structure of the 1st SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide (aa 378-393) from human RIN3 | | Descriptor: | CD2-associated protein, Ras and Rab interactor 3, SULFATE ION | | Authors: | Rouka, E, Simister, P.C, Janning, M, Kirsch, K.H, Krojer, T, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-04 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
3LTF
 
 | | Crystal Structure of the Drosophila Epidermal Growth Factor Receptor ectodomain in complex with Spitz | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Alvarado, D, Klein, D.E, Lemmon, M.A. | | Deposit date: | 2010-02-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for negative cooperativity in growth factor binding to an EGF receptor.
Cell(Cambridge,Mass.), 142, 2010
|
|
2GY7
 
 | | Angiopoietin-2/Tie2 Complex Crystal Structure | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barton, W.A, Nikolov, D.B. | | Deposit date: | 2006-05-09 | | Release date: | 2006-06-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structures of the Tie2 receptor ectodomain and the angiopoietin-2-Tie2 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4FCB
 
 | | Potent and Selective Phosphodiesterase 10A Inhibitors | | Descriptor: | 3,4-dimethyl-1-propyl-7-(quinolin-2-ylmethoxy)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Parris, K.D. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel triazines as potent and selective phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4FFT
 
 | | Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with mixed-linkage glucan | | Descriptor: | ACETIC ACID, Expansin-yoaJ, SULFATE ION, ... | | Authors: | Georgelis, N, Yennawar, N.H, Cosgrove, D.J. | | Deposit date: | 2012-06-01 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8F9H
 
 | | H64A swMb-MeNO adduct | | Descriptor: | Myoglobin, NITROSOMETHANE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herrera, V.E, Thomas, L.M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into Nitrosoalkane Binding to Myoglobin Provided by Crystallography of Wild-Type and Distal Pocket Mutant Derivatives.
Biochemistry, 62, 2023
|
|
8F9I
 
 | | H64A swMb-EtNO adduct | | Descriptor: | Myoglobin, NITROSOETHANE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herrera, V.E, Thomas, L.N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into Nitrosoalkane Binding to Myoglobin Provided by Crystallography of Wild-Type and Distal Pocket Mutant Derivatives.
Biochemistry, 62, 2023
|
|
8F9J
 
 | | H64A swMb-PrNO adduct | | Descriptor: | 1-nitrosopropane, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herrera, V.E, Thomas, L.M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into Nitrosoalkane Binding to Myoglobin Provided by Crystallography of Wild-Type and Distal Pocket Mutant Derivatives.
Biochemistry, 62, 2023
|
|
2JKS
 
 | | Crystal structure of the the bradyzoite specific antigen BSR4 from toxoplasma gondii. | | Descriptor: | BRADYZOITE SURFACE ANTIGEN BSR4, ZINC ION | | Authors: | Boulanger, M.J, Grigg, M.E, Bruic, E, Grujic, O. | | Deposit date: | 2008-08-29 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of the Bradyzoite Surface Antigen (Bsr4) from Toxoplasma Gondii, a Unique Addition to the Surface Antigen Glycoprotein 1-Related Superfamily.
J.Biol.Chem., 284, 2009
|
|
3IEW
 
 | |
