5UAS
 
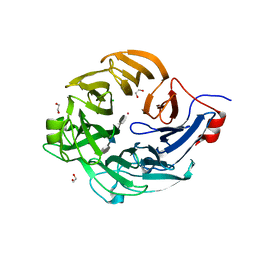 | | Structure of a new family of Polysaccharide lyase PL25-Ulvanlyase bound to -[GlcA(1-4)Rha3S]- | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CHLORIDE ION, ... | | Authors: | Ulaganathan, T.S, Cygler, M. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Ulvan-Degrading Polysaccharide Lyase Family: Structure and Catalytic Mechanism Suggests Convergent Evolution of Active Site Architecture.
ACS Chem. Biol., 12, 2017
|
|
1Z8A
 
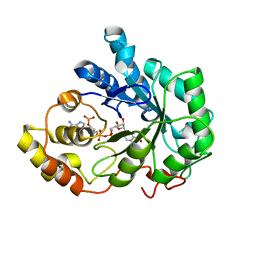 | | Human Aldose Reductase complexed with novel Sulfonyl-pyridazinone Inhibitor | | Descriptor: | 6-[(5-CHLORO-3-METHYL-1-BENZOFURAN-2-YL)SULFONYL]PYRIDAZIN-3(2H)-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Podjarny, A, Heine, A, Klebe, G. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution crystal structure of aldose reductase complexed with the novel sulfonyl-pyridazinone inhibitor exhibiting an alternative active site anchoring group.
J.Mol.Biol., 356, 2006
|
|
7VVR
 
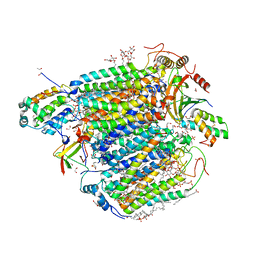 | | Bovine cytochrome c oxidese in CN-bound mixed valence state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
7VUW
 
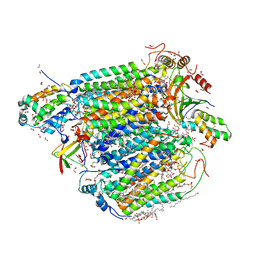 | | Bovine heart cytochrome c oxidase in the cyanide-bound fully oxidized state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
1Z89
 
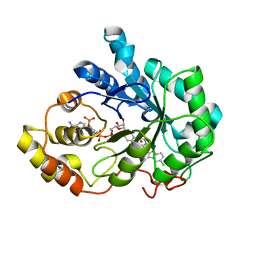 | | Human Aldose Reductase complexed with novel Sulfonyl-pyridazinone Inhibitor | | Descriptor: | 6-[(5-CHLORO-3-METHYL-1-BENZOFURAN-2-YL)SULFONYL]PYRIDAZIN-3(2H)-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Podjarny, A, Heine, A, Klebe, G. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | High-resolution crystal structure of aldose reductase complexed with the novel sulfonyl-pyridazinone inhibitor exhibiting an alternative active site anchoring group.
J.Mol.Biol., 356, 2006
|
|
3NVV
 
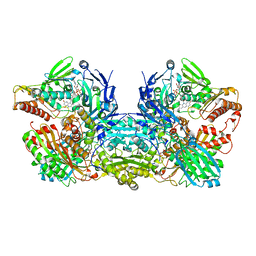 | | Crystal Structure of Bovine Xanthine Oxidase in Complex with Arsenite | | Descriptor: | ARSENITE, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Cao, H, Hille, R. | | Deposit date: | 2010-07-08 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | X-ray Crystal Structure of Arsenite-Inhibited Xanthine Oxidase: Mu-Sulfido,Mu-Oxo Double Bridge between Molybdenum and Arsenic in the Active Site.
J.Am.Chem.Soc., 133, 2011
|
|
3MHU
 
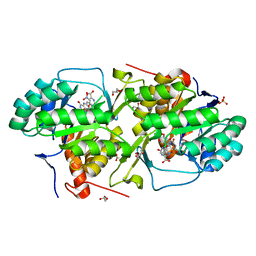 | | Crystal structure of dihydroorotate dehydrogenase from Leishmania major in complex with 5-Nitroorotic acid | | Descriptor: | 5-nitro-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Rocha, J.R, Cheleski, J, Montanari, C.A, Nonato, M.C. | | Deposit date: | 2010-04-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel insights for dihydroorotate dehydrogenase class 1A inhibitors discovery.
Eur.J.Med.Chem., 45, 2010
|
|
3VML
 
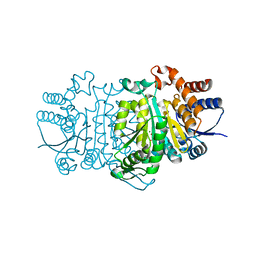 | |
1T2E
 
 | | Plasmodium falciparum lactate dehydrogenase S245A, A327P mutant complexed with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-lactate dehydrogenase, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
3CD5
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[3-(biphenyl-4-ylcarbamoyl)-2-ethyl-5,6,7,8-tetrahydrocyclohepta[b]pyrrol-1(4H)-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
2FZI
 
 | | New Insights into DHFR Interactions: Analysis of Pneumocystis carinii and Mouse DHFR Complexes with NADPH and Two Highly Potent Trimethoprim Derivatives | | Descriptor: | 2,4-DIAMINO-5-[3',4'-DIMETHOXY-5'-(5-CARBOXYL-1-PENTYNYL)]BENZYL PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Chisum, K, Rosowsky, A. | | Deposit date: | 2006-02-09 | | Release date: | 2006-12-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent 5-(omega-carboxy(alkyloxy) trimethoprim derivatives reveals conformational correlations with activity and novel parallel ring stacking interactions.
Proteins, 65, 2006
|
|
1PU8
 
 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 1,N6-ethenoadenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 3H-IMIDAZO[2,1-I]PURINE, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
5OWW
 
 | | Crystal structure of human BRD4(1) bromodomain in complex with UT22B | | Descriptor: | Bromodomain-containing protein 4, ~{N}-(3-methylbenzotriazol-5-yl)-1-(phenylmethyl)imidazole-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-09-04 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple).
Acs Cent.Sci., 4, 2018
|
|
2XAP
 
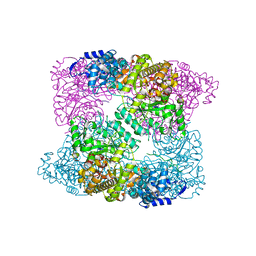 | |
1FO4
 
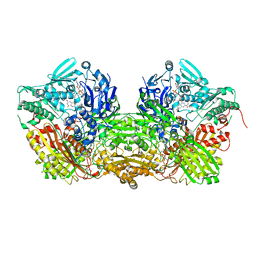 | | CRYSTAL STRUCTURE OF XANTHINE DEHYDROGENASE ISOLATED FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4GX0
 
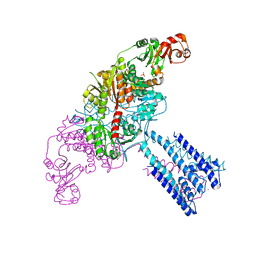 | | Crystal structure of the GsuK L97D mutant | | Descriptor: | CALCIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | Deposit date: | 2012-09-03 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
2A92
 
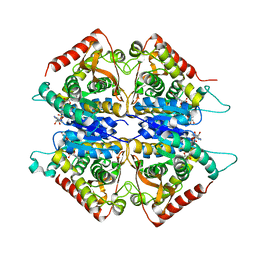 | | Crystal structure of lactate dehydrogenase from Plasmodium vivax: complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase | | Authors: | Chaikuad, A, Fairweather, V, Conners, R, Joseph-Horne, T, Turgut-Balik, D, Brady, R.L. | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of Lactate Dehydrogenase from Plasmodium vivax: Complexes with NADH and APADH.
Biochemistry, 44, 2005
|
|
1BG9
 
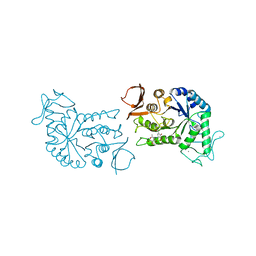 | | BARLEY ALPHA-AMYLASE WITH SUBSTRATE ANALOGUE ACARBOSE | | Descriptor: | 1,4-ALPHA-D-GLUCAN GLUCANOHYDROLASE, 4,6-dideoxy-4-{[(1S,4S,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Kadziola, A, Haser, R. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular structure of a barley alpha-amylase-inhibitor complex: implications for starch binding and catalysis.
J.Mol.Biol., 278, 1998
|
|
4K9U
 
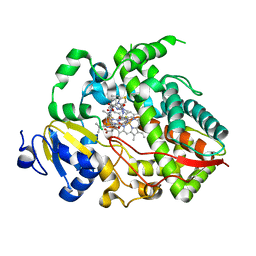 | | Complex of human CYP3A4 with a desoxyritonavir analog | | Descriptor: | Cytochrome P450 3A4, N~2~-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-N-[(2S,5S)-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}hexan-2-yl]-D-valinamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dissecting Cytochrome P450 3A4-Ligand Interactions Using Ritonavir Analogues.
Biochemistry, 52, 2013
|
|
2FZJ
 
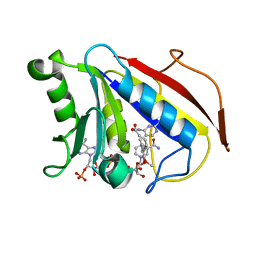 | | New Insights into DHFR Interactions: Analysis of Pneumocystis carinii and Mouse DHFR Complexes with NADPH and Two Highly Potent Trimethoprim Derivatives | | Descriptor: | 2,4-DIAMINO-5-[3',4'-DIMETHOXY-5'-(5-CARBOXYL-1-PENTYNYL)]BENZYL PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Chisum, K, Rosowsky, A. | | Deposit date: | 2006-02-09 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent 5-(omega-carboxy(alkyloxy) trimethoprim derivatives reveals conformational correlations with activity and novel parallel ring stacking interactions.
Proteins, 65, 2006
|
|
3NXO
 
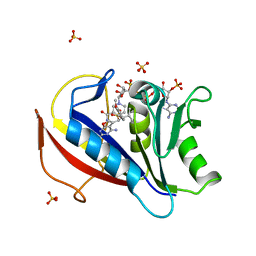 | | Perferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1Z)-2-(2-methoxyphenyl)-3-methylbut-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7M2O
 
 | | Crystal structure of Human Glycolate Oxidase with Inhibitor Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(5'-{1-(4-carboxy-1,3-thiazol-2-yl)-5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-1H-pyrazol-3-yl}-2'-fluoro[1,1'-biphenyl]-4-yl)oxy]-1H-1,2,3-triazole-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gumpena, R, Ding, J, Powell, D.A, Lowther, W.T. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Dual Glycolate Oxidase/Lactate Dehydrogenase A Inhibitors for Primary Hyperoxaluria
ACS Medicinal Chemistry Letters, 12, 2021
|
|
3O5A
 
 | | Crystal Structure of partially reduced Periplasmic Nitrate Reductase from Cupriavidus necator using Ionic Liquids | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CHLORIDE ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Coelho, C, Trincao, J, Romao, M.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states
J.Mol.Biol., 408, 2011
|
|
4EKS
 
 | | T4 Lysozyme L99A/M102H with Isoxazole Bound | | Descriptor: | 1,2-benzisoxazole, 2-HYDROXYETHYL DISULFIDE, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Engineering a model protein cavity to catalyze the Kemp elimination.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5P8W
 
 | | Crystal Structure of COMT in complex with [5-(2,4-dimethyl-1,3-thiazol-5-yl)-1H-pyrazol-3-yl]methanamine | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Catechol O-methyltransferase, FORMIC ACID, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-25 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of COMT complex
To be published
|
|
