2XOE
 
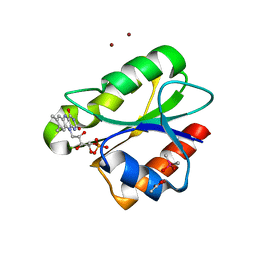 | | Crystal structure of flavoprotein NrdI from Bacillus anthracis in the semiquinone form | | Descriptor: | ACETATE ION, CACODYLATE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Johansson, R, Sprenger, J, Torrents, E, Sahlin, M, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2010-08-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Crystal Structures of Nrdi in the Oxidised and Reduced States: An Unusual Flavodoxin
FEBS J., 277, 2010
|
|
8V35
 
 | | Crystal structure of HpsN from Cupriavidus pinatubonensis | | Descriptor: | 1,2-ETHANEDIOL, Sulfopropanediol 3-dehydrogenase, ZINC ION | | Authors: | Lee, M. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and kinetic insights into the stereospecific oxidation of R -2,3-dihydroxypropanesulfonate by DHPS-3-dehydrogenase from Cupriavidus pinatubonensis.
Chem Sci, 15, 2024
|
|
8V37
 
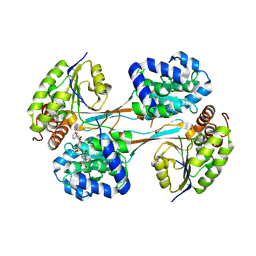 | |
8V36
 
 | |
4EKX
 
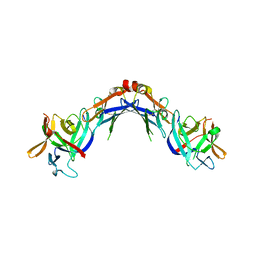 | |
2VNC
 
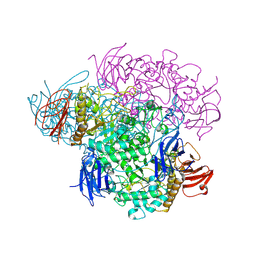 | | Crystal structure of Glycogen Debranching enzyme TreX from Sulfolobus solfataricus | | Descriptor: | GLYCOGEN OPERON PROTEIN GLGX | | Authors: | Song, H.-N, Yoon, S.-M, Cha, H, Park, K.-T, Woo, E.-J. | | Deposit date: | 2008-02-04 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
7AL7
 
 | | The Crystal Structure of Human IL-18 in Complex With Human IL-18 Binding Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutathione S-transferase class-mu 26 kDa isozyme,Interleukin-18, Interleukin-18-binding protein | | Authors: | Detry, S, Andries, J, Bloch, Y, Clancy, D, Savvides, S.N. | | Deposit date: | 2020-10-05 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of human IL-18 sequestration by the decoy receptor IL-18 binding protein in inflammation and tumor immunity.
J.Biol.Chem., 298, 2022
|
|
2VEO
 
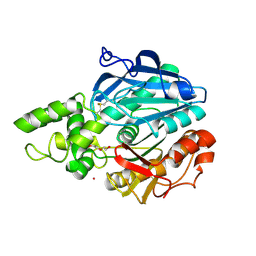 | | X-ray structure of Candida antarctica lipase A in its closed state. | | Descriptor: | GLYCEROL, LIPASE A, TETRAETHYLENE GLYCOL, ... | | Authors: | Ericsson, D.J, Kasrayan, A, Johansson, P, Bergfors, T, Sandstrom, A.G, Backvall, J.E, Mowbray, S.L. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structure of Candida Antarctica Lipase a Shows a Novel Lid Structure and a Likely Mode of Interfacial Activation.
J.Mol.Biol., 376, 2008
|
|
7E35
 
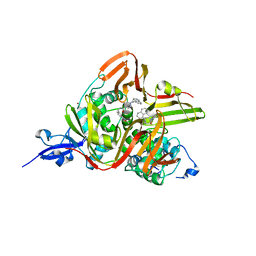 | | Crystal structure of the SARS-CoV-2 papain-like protease (PLPro) C112S mutant bound to compound S43 | | Descriptor: | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Non-structural protein 3, ZINC ION | | Authors: | Liu, J, Wang, Y, Xu, X, Pan, L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2.
Cell Chem Biol, 28, 2021
|
|
4DKP
 
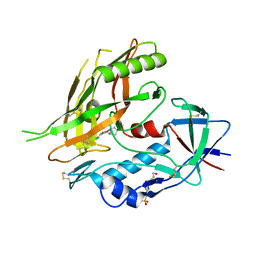 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with AWS-I-50 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-[(1S,2S)-2-amino-2,3-dihydro-1H-inden-1-yl]-N'-(4-chloro-3-fluorophenyl)ethanediamide, ... | | Authors: | Kwon, Y.D, LaLonde, J.M, Jones, D.M, Sun, A.W, Courter, J.R, Soeta, T, Kobayashi, T, Princiotto, A.M, Wu, X, Mascola, J, Schon, A, Freire, E, Sodroski, J, Madani, N, Smith III, A.B, Kwong, P.D. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7978 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of Dual Hotspot Small-Molecule HIV-1 Entry Inhibitors.
J.Med.Chem., 55, 2012
|
|
2XYQ
 
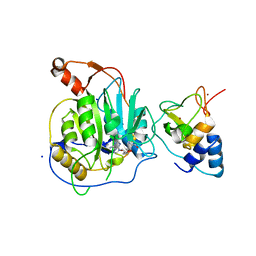 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
7BUJ
 
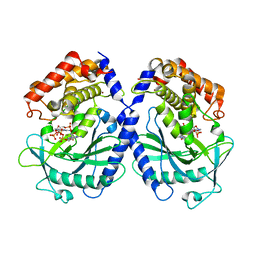 | | mcGAS bound with pppGpG | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, B, Su, X.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Mn2+Directly Activates cGAS and Structural Analysis Suggests Mn2+Induces a Noncanonical Catalytic Synthesis of 2'3'-cGAMP.
Cell Rep, 32, 2020
|
|
2Y1M
 
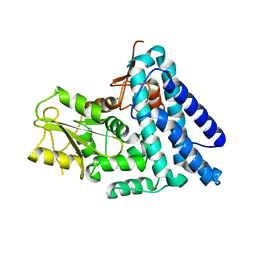 | | Structure of native c-Cbl | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE, ZINC ION | | Authors: | Dou, H, Sibbet, G.J, Huang, D.T. | | Deposit date: | 2010-12-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7DTP
 
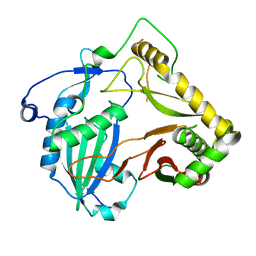 | | Crystal structure of agmatine coumaroyltransferase from Triticum aestivum | | Descriptor: | agmatine coumaroyltransferase | | Authors: | Yamane, M, Takenoya, M, Sue, M, Yajima, S. | | Deposit date: | 2021-01-06 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular and structural characterization of agmatine coumaroyltransferase in Triticeae, the key regulator of hydroxycinnamic acid amide accumulation.
Phytochemistry, 189, 2021
|
|
4DKR
 
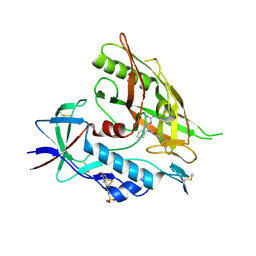 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with AWS-I-169 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core, ... | | Authors: | Kwon, Y.D, LaLonde, J.M, Jones, D.M, Sun, A.W, Courter, J.R, Soeta, T, Kobayashi, T, Princiotto, A.M, Wu, X, Mascola, J, Schon, A, Freire, E, Sodroski, J, Madani, N, Smith III, A.B, Kwong, P.D. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of Dual Hotspot Small-Molecule HIV-1 Entry Inhibitors.
J.Med.Chem., 55, 2012
|
|
7T7N
 
 | | Structure of SPCC1393.13 protein from fission yeast | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Damage-control phosphatase SPCC1393.13, PHOSPHATE ION | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
7T7O
 
 | |
7T7K
 
 | | Structure of SPAC806.04c protein from fission yeast bound to Co2+ | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Damage-control phosphatase SPAC806.04c, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
7ESC
 
 | | FmnB complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-05-10 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7ESA
 
 | | the complex structure of flavin transferase FmnB complexed with FAD | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
6KLK
 
 | |
7ESB
 
 | | FmnB complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7F39
 
 | | The structure of flavin transferase FmnB | | Descriptor: | FAD:protein FMN transferase | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-06-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7F2U
 
 | | FmnB complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-06-14 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7TZJ
 
 | | SARS CoV-2 PLpro in complex with inhibitor 3k | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3-fluorophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Papain-like protease, ... | | Authors: | Calleja, D.J, Klemm, T, Lechtenberg, B.C, Kuchel, N.W, Lessene, G, Komander, D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insights Into Drug Repurposing, as Well as Specificity and Compound Properties of Piperidine-Based SARS-CoV-2 PLpro Inhibitors.
Front Chem, 10, 2022
|
|
