5F9O
 
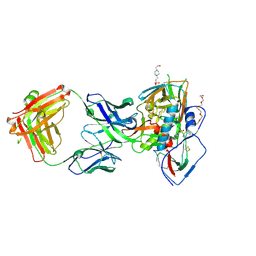 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235.09 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CH235.09 Light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
5M7M
 
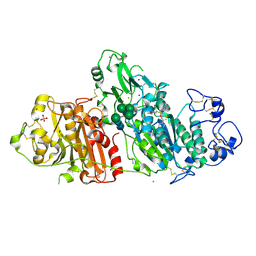 | | Novel Imidazo[1,2-a]pyridine Derivatives with Potent Autotaxin/ENPP2 Inhibitor Activity | | Descriptor: | CHLORIDE ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, IODIDE ION, ... | | Authors: | Wolhkoning, A, Fleury, D, Leonard, P, Triballeau, N, Mollat, P, Vercheval, L. | | Deposit date: | 2016-10-28 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, Structure-Activity Relationship, and Binding Mode of an Imidazo[1,2-a]pyridine Series of Autotaxin Inhibitors.
J. Med. Chem., 60, 2017
|
|
4LKV
 
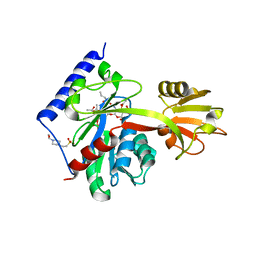 | | Determinants of lipid substrate and membrane binding for the tetraacyldisaccharide-1-phosphate 4 -kinase LpxK | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Emptage, R.P, Tonthat, N.K, York, J.D, Schumacher, M.A, Zhou, P. | | Deposit date: | 2013-07-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5109 Å) | | Cite: | Structural Basis of Lipid Binding for the Membrane-embedded Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK.
J.Biol.Chem., 289, 2014
|
|
3EE5
 
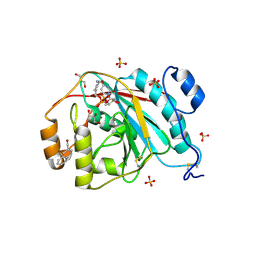 | | Crystal structure of human M340H-Beta1,4-Galactosyltransferase-I (M340H-B4GAL-T1) in complex with GLCNAC-Beta1,3-Gal-Beta-Naphthalenemethanol | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2008-09-04 | | Release date: | 2009-01-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Deoxygenated Disaccharide Analogs as Specific Inhibitors of {beta}1-4-Galactosyltransferase 1 and Selectin-mediated Tumor Metastasis
J.Biol.Chem., 284, 2009
|
|
6DV0
 
 | | HIV-1 wild type protease with GRL-02815A, a thiochroman heterocycle with (S)-Boc-amine functionality as the P2 ligand | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Design, synthesis, and X-ray studies of potent HIV-1 protease inhibitors incorporating aminothiochromane and aminotetrahydronaphthalene carboxamide derivatives as the P2 ligands.
Eur J Med Chem, 160, 2018
|
|
3E7B
 
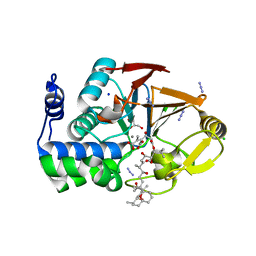 | | Crystal Structure of Protein Phosphatase-1 Bound to the natural toxin inhibitor Tautomycin | | Descriptor: | (2Z)-2-[(1R)-3-{[(1R,2S,3R,6S,7S,10R)-10-{(2S,3S,6R,8S,9R)-3,9-dimethyl-8-[(3S)-3-methyl-4-oxopentyl]-1,7-dioxaspiro[5.5]undec-2-yl}-3,7-dihydroxy-2-methoxy-6-methyl-1-(1-methylethyl)-5-oxoundecyl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, AZIDE ION, CHLORIDE ION, ... | | Authors: | Kelker, M.S, Page, R, Peti, W. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors
J.Mol.Biol., 385, 2009
|
|
2WMS
 
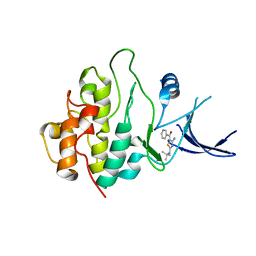 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE CHK1, [4-amino-2-(tert-butylamino)-1,3-thiazol-5-yl](phenyl)methanone | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
5ZZV
 
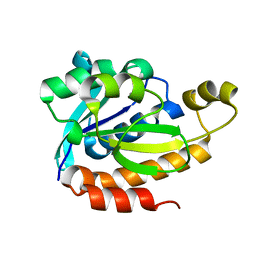 | | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | Authors: | Bairagya, H.R, Sharma, P, Iqbal, N, Sharma, S, Singh, T.P. | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution
To Be Published
|
|
2X39
 
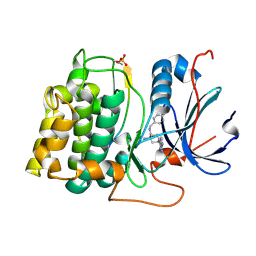 | | Structure of 4-Amino-N-(4-chlorobenzyl)-1-(7H-pyrrolo(2,3-d)pyrimidin- 4-yl)piperidine-4-carboxamide bound to PKB | | Descriptor: | 4-AMINO-N-(4-CHLOROBENZYL)-1-(7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)PIPERIDINE-4-CARBOXAMIDE, GLYCOGEN SYNTHASE KINASE-3 BETA, RAC-BETA SERINE/THREONINE-PROTEIN KINASE | | Authors: | Davies, T.G, McHardy, T, Caldwell, J.J, Cheung, K.M, Hunter, L.J, Taylor, K, Rowlands, M, Ruddle, R, Henley, A, Brandon, A.D, Valenti, M, Fazal, L, Seavers, L, Raynaud, F.I, Eccles, S.A, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of 4-Amino-1-(7H-Pyrrolo[2,3-D]Pyrimidin-4-Yl)Piperidine-4-Carboxamides as Selective, Orally Active Inhibitors of Protein Kinase B (Akt).
J.Med.Chem., 53, 2010
|
|
5LP1
 
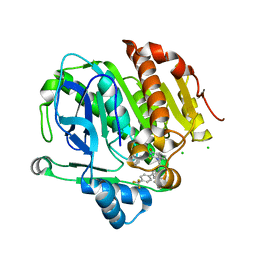 | |
6DIL
 
 | | HIV-1 protease with single mutation L76V in complex with tipranavir | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wong-Sam, A.E, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2018-05-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
6DJ2
 
 | | HIV-1 protease with single mutation L76V in complex with Lopinavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Zhang, Y, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
6DJ1
 
 | | Wild-type HIV-1 protease in complex with Lopinavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Zhang, Y, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
5DGW
 
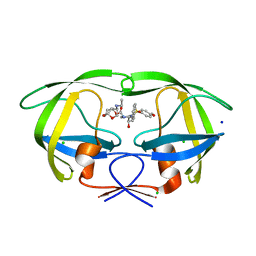 | | Crystal Structure of HIV-1 Protease Inhibitor GRL-105-11A Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand | | Descriptor: | (3R,3aS,4S,7aS)-3-(ethylamino)hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Pol protein, ... | | Authors: | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2015-08-28 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
6F1J
 
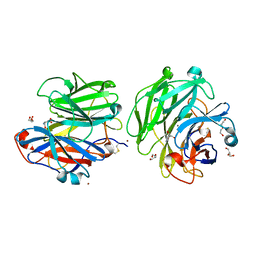 | | Structure of a Talaromyces pinophilus GH62 Arabinofuranosidase in complex with AraDNJ at 1.25A resolution | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Sobala, L, Blagova, E, Coyle, T, Morkeberg Krogh, K.B.R, Wei, P, Stubbs, K, Wilson, K.S, Davies, G.J. | | Deposit date: | 2017-11-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of a Talaromyces pinophilus GH62 arabinofuranosidase in complex with AraDNJ at 1.25 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3EQN
 
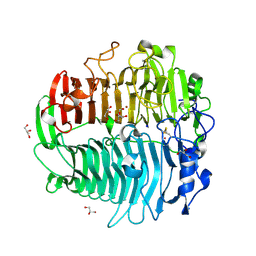 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) | | Descriptor: | ACETATE ION, GLYCEROL, Glucan 1,3-beta-glucosidase, ... | | Authors: | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
4ZBI
 
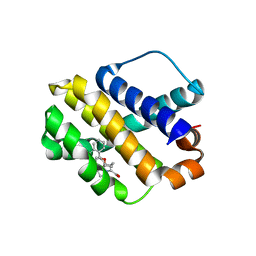 | | Mcl-1 complexed with small molecules | | Descriptor: | 1-[3-(naphthalen-1-yloxy)propyl]-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of tricyclic indoles that potently inhibit mcl-1 using fragment-based methods and structure-based design.
J.Med.Chem., 58, 2015
|
|
3EQO
 
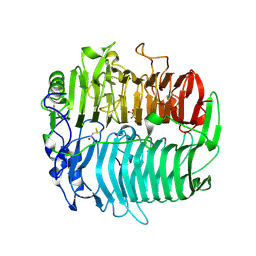 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) gluconolactone complex | | Descriptor: | D-glucono-1,5-lactone, Glucan 1,3-beta-glucosidase, ZINC ION, ... | | Authors: | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
8D0X
 
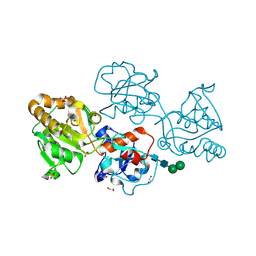 | | Human FUT9 bound to LNnT | | Descriptor: | 1,2-ETHANEDIOL, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
8D0S
 
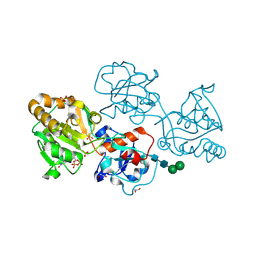 | | Human FUT9 bound to GDP and LNnT | | Descriptor: | 1,2-ETHANEDIOL, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
5DGU
 
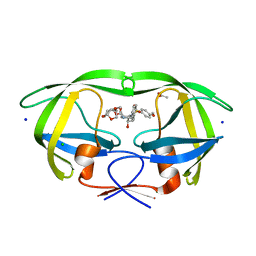 | | Crystal Structure of HIV-1 Protease Inhibitors Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand GRL-004-11A | | Descriptor: | (3R,3aR,4S,7aS)-3-methoxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2015-08-28 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
4ZAC
 
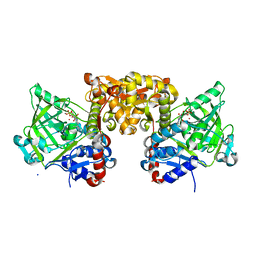 | | Structure of S. cerevisiae Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
5EOL
 
 | |
7LPX
 
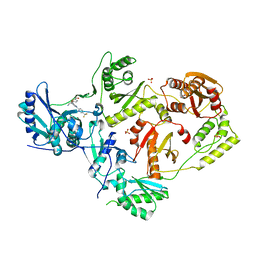 | | Crystal Structure of HIV-1 RT in Complex with NBD-14270 | | Descriptor: | Reverse transcriptase p51, Reverse transcriptase p66, SULFATE ION, ... | | Authors: | Losada, N, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-12 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J.Med.Chem., 64, 2021
|
|
7LQU
 
 | | Crystal Structure of HIV-1 RT in Complex with NBD-14075 | | Descriptor: | Reverse transcriptase p51, Reverse transcriptase p66, SULFATE ION, ... | | Authors: | Losada, N, Ruiz, F.X, Gruber, K, Das, K, Arnold, E. | | Deposit date: | 2021-02-15 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J.Med.Chem., 64, 2021
|
|
