6ECE
 
 | | Vlm2 thioesterase domain with genetically encoded 2,3-diaminopropionic acid bound with a dodecadepsipeptide, space group H3 | | Descriptor: | Vlm2, dodecadepsipeptide | | Authors: | Alonzo, D.A, Huguenin-Dezot, N, Heberlig, G.W, Mahesh, M, Nguyen, D.P, Dornan, M.H, Boddy, C.N, Chin, J.W, Schmeing, T.M. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid.
Nature, 565, 2019
|
|
8QHQ
 
 | | Crystal structure of human DNPH1 bound to hmdUMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
7K6F
 
 | | Crystal structure of the tandem bromodomain (BD1, BD2) of human TAF1 in complex with MES (2-(N-morpholino)ethanesulfonic acid) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2020-09-20 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
6IOU
 
 | | The ligand binding domain of Mlp24 with serine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6AZN
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
8QLQ
 
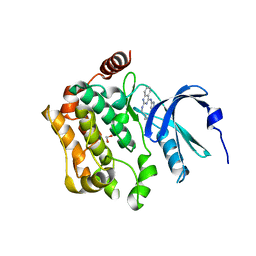 | | Human MST3 (STK24) kinase in complex with macrocyclic inhibitor JA310 | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase 24, macrocyclic inhibitor | | Authors: | Balourdas, D.I, Amrhein, J.A, Hanke, T, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Synthesis of Pyrazole-Based Macrocycles Leads to a Highly Selective Inhibitor for MST3.
J.Med.Chem., 67, 2024
|
|
6XK3
 
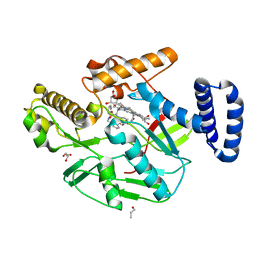 | |
8A3I
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil homotetramer with 3 heptad repeats, apCC-Tet*3 | | Descriptor: | apCC-Tet*3 | | Authors: | Naudin, E.A, Mylemans, B, Albanese, K.I, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
6IRX
 
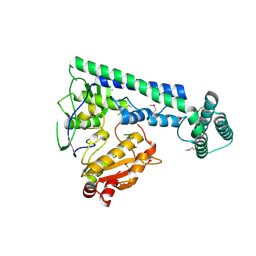 | |
8QLS
 
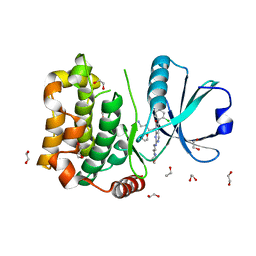 | | Human MST3 (STK24) kinase in complex with inhibitor MR26 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-(3-morpholin-4-ylpropylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
6P78
 
 | | queuine lyase from Clostridium spiroforme bound to SAM and queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5S)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, IRON/SULFUR CLUSTER, Queuine lyase, ... | | Authors: | Almo, S.C, Grove, T.L. | | Deposit date: | 2019-06-05 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | Discovery of novel bacterial queuine salvage enzymes and pathways in human pathogens.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6EE6
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6o) and catalytic zinc ion | | Descriptor: | (2R)-2-[(cyclopentylacetyl)amino]-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
8A3K
 
 | | X-ray crystal structure of a de novo designed single-chain antiparallel 4-helix coiled-coil bundle, sc-apCC-4 | | Descriptor: | sc-apCC-4 | | Authors: | Albanese, K.I, Mylemans, B, Naudin, E.A, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
6EEI
 
 | | Crystal structure of Arabidopsis thaliana phenylacetaldehyde synthase in complex with L-phenylalanine | | Descriptor: | PHENYLALANINE, SULFATE ION, Tyrosine decarboxylase 1 | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-14 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (1.99001348 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XK6
 
 | |
8QHR
 
 | | Crystal structure of the human DNPH1 glycosyl-enzyme intermediate | | Descriptor: | 1',2'-DIDEOXYRIBOFURANOSE-5'-PHOSPHATE, 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, ... | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
7ZV2
 
 | | HUMAN PRMT5:MEP50 Crystal Structure With MTA and Fragment Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, Methylosome protein 50, ... | | Authors: | Ahmad, M.U, Koelmel, W, Arkhipova, V, Lawson, J.D, Smith, C.R, Gunn, R.J. | | Deposit date: | 2022-05-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7ZZN
 
 | |
6P6S
 
 | | HCV NS3/4A protease domain of genotype 3a in complex with glecaprevir | | Descriptor: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, 1,2-ETHANEDIOL, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of pan-genotypic HCV NS3/4A protease inhibition by glecaprevir and characterization of genotype-specific structural differences
To Be Published
|
|
6ISV
 
 | | Structure of acetophenone reductase from Geotrichum candidum NBRC 4597 in complex with NAD | | Descriptor: | Acetophenone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Koesoema, A.A, Sugiyama, Y, Senda, M, Senda, T, Matsuda, T. | | Deposit date: | 2018-11-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for a highly (S)-enantioselective reductase towards aliphatic ketones with only one carbon difference between side chain.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
6EOY
 
 | | Transthyretin in complex with 4-(1,3-Benzothiazol-2-yl)-2-methylaniline | | Descriptor: | 4-(1,3-benzothiazol-2-yl)-2-methyl-aniline, Transthyretin | | Authors: | Leite, J.P, Gales, L. | | Deposit date: | 2017-10-10 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting transthyretin in Alzheimer's disease: Drug discovery of small-molecule chaperones as disease-modifying drug candidates for Alzheimer's disease.
Eur.J.Med.Chem., 226, 2021
|
|
5EVF
 
 | | Crystal structure of a Francisella virulence factor FvfA in the hexagonal form | | Descriptor: | CHLORIDE ION, Francisella virulence factor, GLYCEROL | | Authors: | Kolappan, S, Lo, K.Y, Shen, C.L.J, Guttman, J.A, Craig, L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-10-26 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structure of the conserved Francisella virulence protein FvfA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6P6V
 
 | | HCV NS3/4A protease domain of genotype 5a in complex with glecaprevir | | Descriptor: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, 1,2-ETHANEDIOL, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of pan-genotypic HCV NS3/4A protease inhibition by glecaprevir and characterization of genotype-specific structural differences
To Be Published
|
|
7ZM1
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclophostin-like inhibitor CyC7b | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, methoxy-[(~{E},3~{R})-3-[(2~{R})-1-methoxy-1,3-bis(oxidanylidene)butan-2-yl]tridec-11-enyl]phosphinous acid | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
6BDN
 
 | | Crystal structure of human TAO3 kinase binding ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Aleshin, A.E, Bankton, L.A, Pinkerton, A, Courtneidge, S.A, Liddington, R.C. | | Deposit date: | 2017-10-23 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human TAO3 kinase binding ADP
To Be Published
|
|
