6CJY
 
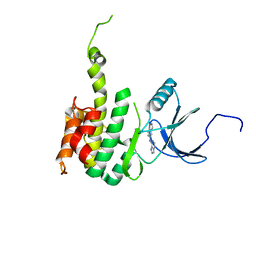 | | Crystal Structure of Mnk2-D228G in complex with Inhibitor | | Descriptor: | 5-[(7H-purin-6-yl)amino]-1H-isoindol-1-one, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6CJW
 
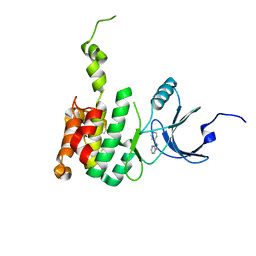 | | Crystal Structure of Mnk2-D228G in Complex With Inhibitor | | Descriptor: | 3-(pyridin-4-yl)imidazo[1,2-b]pyridazine, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
7CX4
 
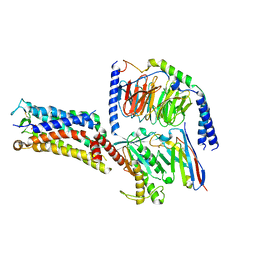 | | Cryo-EM structure of the Evatanepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-~{tert}-butylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX3
 
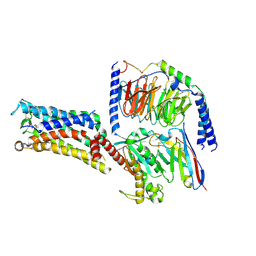 | | Cryo-EM structure of the Taprenepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-pyrazol-1-ylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7H6Z
 
 | | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1272494722 (CHIKV_MacB-x0406) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Dolci, I, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Oliva, G, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Group deposition for crystallographic fragment screening of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
7WI3
 
 | | Cryo-EM structure of E.Coli FtsH-HflkC AAA protease complex | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, Modulator of FtsH protease HflC, Modulator of FtsH protease HflK | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2022-01-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the entire FtsH-HflKC AAA protease complex.
Cell Rep, 39, 2022
|
|
7MBV
 
 | | Cryo-EM structure of zebrafish TRPM5 in the presence of 5 mM calcium and 0.5 mM NDNA | | Descriptor: | (25R)-14beta,17beta-spirost-5-en-3beta-ol, (2R)-2-(hydroxymethyl)-4-{[(25R)-10alpha,14beta,17beta-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruan, Z, Lu, W, Du, J, Haley, E. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of the TRPM5 channel elucidate mechanisms of activation and inhibition.
Nat.Struct.Mol.Biol., 28, 2021
|
|
9D0U
 
 | | Crystal structure of CDK2 in complex with Cpd 2 | | Descriptor: | 6-chloro-8-cyclopentyl-2-[4-(ethanesulfonyl)-2-methylanilino]pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2 | | Authors: | Kwiatkowski, N, Liang, T, Sha, Z, Collier, P.N, Yang, A, Sathappa, M, Paul, A, Su, L, Zheng, X, Aversa, R, Li, K, Mehovic, R, Breitkopf, S.B, Chen, D, Howarth, C.L, Yuan, K, Jo, H, Growney, J.D, Weiss, M, Williams, J. | | Deposit date: | 2024-08-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CDK2 heterobifunctional degraders co-degrade CDK2 and cyclin E resulting in efficacy in CCNE1-amplified and overexpressed cancers.
Cell Chem Biol, 32, 2025
|
|
8UY6
 
 | | Aquaporin Z with ALFA tag and bound to nanobody | | Descriptor: | Aquaporin Z, CARDIOLIPIN, anti-ALFA nanobody | | Authors: | Stover, L, Bahramimoghaddam, H, Wang, L, Zhou, M, Laganowsky, A. | | Deposit date: | 2023-11-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Grafting the ALFA tag for structural studies of aquaporin Z.
J Struct Biol X, 9, 2024
|
|
8GT6
 
 | | human STING With agonist HB3089 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
7Z59
 
 | | SARS-CoV-2 main protease (Mpro) covalently modified with a penicillin derivative | | Descriptor: | (3S)-4-[[2,4-bis(fluoranyl)phenyl]methoxy]-2-methyl-4-oxidanylidene-3-[[(Z)-3-oxidanylidene-2-(2-phenoxyethanoylamino)prop-1-enyl]amino]butane-2-sulfinic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5 | | Authors: | Owen, C.D, Malla, T.R, Brewitz, L, Lukacik, P, Strain-Damerell, C, Mikolajek, H, Muntean, D.G, Aslam, H, Salah, E, Tumber, A, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-03-08 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Penicillin Derivatives Inhibit the SARS-CoV-2 Main Protease by Reaction with Its Nucleophilic Cysteine.
J.Med.Chem., 65, 2022
|
|
3PKX
 
 | | Crystal Structure of Toxoflavin Lyase (TflA) bound to Mn(II) and Toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, MANGANESE (II) ION, Toxoflavin lyase (TflA) | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Toxoflavin lyase requires a novel 1-his-2-carboxylate facial triad .
Biochemistry, 50, 2011
|
|
3FUG
 
 | | Crystal Structure of the Retinoid X Receptor Ligand Binding Domain Bound to the Synthetic Agonist 3-[4-Hydroxy-3-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-phenyl]acrylic Acid | | Descriptor: | (2E)-3-[4-hydroxy-3-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)phenyl]prop-2-enoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Bourguet, W. | | Deposit date: | 2009-01-14 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modulating retinoid X receptor with a series of (E)-3-[4-hydroxy-3-(3-alkoxy-5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)phenyl]acrylic acids and their 4-alkoxy isomers.
J.Med.Chem., 52, 2009
|
|
5VP2
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with madumycin II and bound to mRNA and A-, P- and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Osterman, I.A, Khabibullina, N.F, Komarova, E.S, Kasatsky, P, Kartsev, V.G, Bogdanov, A.A, Dontsova, O.A, Konevega, A.L, Sergiev, P.V, Polikanov, Y.S. | | Deposit date: | 2017-05-04 | | Release date: | 2017-06-28 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Madumycin II inhibits peptide bond formation by forcing the peptidyl transferase center into an inactive state.
Nucleic Acids Res., 45, 2017
|
|
4O24
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{Z})-5-[(4-hydroxyphenyl)methylidene]-3-(2-methylpropyl)-2-sulfanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
5W19
 
 | | Tryptophan indole-lyase complex with oxindolyl-L-alanine | | Descriptor: | 1-carboxy-1-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]azaniumyl}-2-[(3R)-2-oxo-2,3-dihydro-1H-indol-3-yl]ethan-1-ide, POTASSIUM ION, Tryptophanase | | Authors: | Phillips, R.S, Wood, Z.A. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of Proteus vulgaris tryptophan indole-lyase complexed with oxindolyl-L-alanine: implications for the reaction mechanism.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5VQ5
 
 | | Crystal Structure of the Lectin Domain From the F17-like Adhesin, UclD | | Descriptor: | Adhesin, IODIDE ION | | Authors: | Klein, R.D, Spaulding, C.N, Dodson, K.W, Pinkner, J.S, Hultgren, S.J, Fremont, D. | | Deposit date: | 2017-05-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist.
Nature, 546, 2017
|
|
9ECT
 
 | |
6CCF
 
 | | Crystal Structure of the Human CAMKK1A in complex with Hesperadin | | Descriptor: | 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase kinase 1, N-[2-OXO-3-((E)-PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]IMINO}METHYL)-2,6-DIHYDRO-1H-INDOL-5-YL]ETHANESULFONAMIDE, ... | | Authors: | Santiago, A.S, Counago, R.M, dos Reis, C.V, Ramos, P.Z, Silva, P.N.B, Drewry, D, Elkins, J.M, Massirer, K.B, Arruda, P, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human CAMKK1A in complex with Hesperadin
To be Published
|
|
5W4K
 
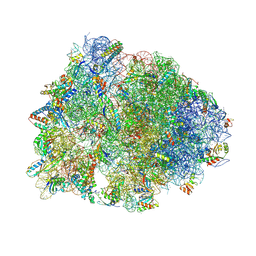 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Klebsazolicin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Metelev, M, Osterman, I.A, Ghilarov, D, Khabibullina, N.F, Yakimov, A, Shabalin, K, Utkina, I, Travin, D.Y, Komarova, E.S, Serebryakova, M, Artamonova, T, Khodorkovskii, M, Konevega, A.L, Sergiev, P.V, Severinov, K, Polikanov, Y.S. | | Deposit date: | 2017-06-12 | | Release date: | 2017-08-30 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel.
Nat. Chem. Biol., 13, 2017
|
|
3GCM
 
 | |
7B9H
 
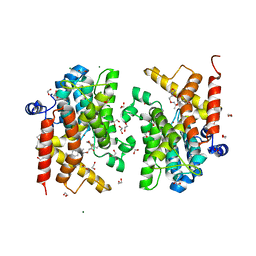 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-42a | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-1-(3-cyclopentyloxy-4-methoxy-phenyl)ethylideneamino]oxy-1-morpholin-4-yl-propan-1-one, MAGNESIUM ION, ... | | Authors: | Torretta, A, Abbate, S, Parisini, E. | | Deposit date: | 2020-12-14 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, biological evaluation and structural characterization of novel GEBR library PDE4D inhibitors.
Eur.J.Med.Chem., 223, 2021
|
|
2JJ1
 
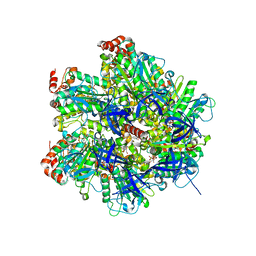 | | The Structure of F1-ATPase inhibited by piceatannol. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1ZL5
 
 | | Crystal structure of Glu335Gln mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | CHLORIDE ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-05 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
5M55
 
 | | Nek2 bound to arylaminopurine 71 | | Descriptor: | 6-[(~{Z})-2-(diethylamino)ethenyl]-~{N}-phenyl-7~{H}-purin-2-amine, CHLORIDE ION, SULFATE ION, ... | | Authors: | Bayliss, R. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided design of purine-based probes for selective Nek2 inhibition.
Oncotarget, 8, 2017
|
|
