7VYL
 
 | |
7VYK
 
 | |
7VYM
 
 | |
7VXZ
 
 | |
7ST5
 
 | | Structure of Fab CC-95251 in complex with SIRP-alpha | | Descriptor: | 1,2-ETHANEDIOL, Fab CC-95251 anti-SIRP-alpha heavy chain, Fab CC-95251 anti-SIRP-alpha light chain, ... | | Authors: | Fenalti, G. | | Deposit date: | 2021-11-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Preclinical Activity of BMS-986351, an Antibody to SIRP alpha That Enhances Macrophage-mediated Tumor Phagocytosis When Combined with Opsonizing Antibodies.
Cancer Res Commun, 4, 2024
|
|
7VUX
 
 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
7VUN
 
 | | Design, modification, evaluation and cocrystal studies of novel phthalimides regulating PD-1/PD-L1 interaction | | Descriptor: | (2~{S},3~{S})-2-[[6-[(3-cyanophenyl)methoxy]-2-(2-methyl-3-phenyl-phenyl)-1,3-bis(oxidanylidene)isoindol-5-yl]methylamino]-3-oxidanyl-butanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Sun, C.L, Chen, M.R, Yang, P, Xiao, Y.B. | | Deposit date: | 2021-11-03 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Novel phthalimides regulating PD-1/PD-L1 interaction as potential immunotherapy agents.
Acta Pharm Sin B, 12, 2022
|
|
7SJQ
 
 | |
7RYN
 
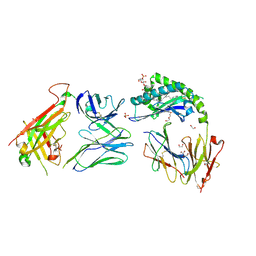 | | CD1a-sulfatide-gdTCR complex | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-08-25 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atypical sideways recognition of CD1a by autoreactive gamma delta T cell receptors.
Nat Commun, 13, 2022
|
|
7RYL
 
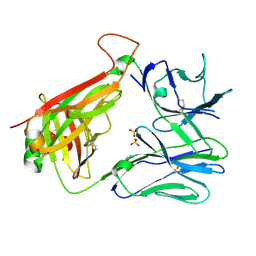 | | T cell receptor CO3 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, T cell receptor delta variable 1,T cell receptor alpha chain constant, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-08-25 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atypical sideways recognition of CD1a by autoreactive gamma delta T cell receptors.
Nat Commun, 13, 2022
|
|
7RYO
 
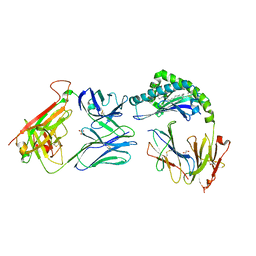 | | CD1a-dideoxymycobactin-gdTCR complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, SULFATE ION, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-08-25 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Atypical sideways recognition of CD1a by autoreactive gamma delta T cell receptors.
Nat Commun, 13, 2022
|
|
7RYM
 
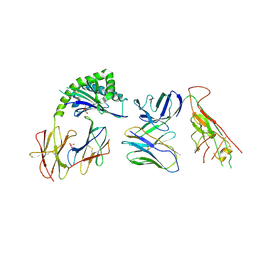 | | CD1a-endo-gdTCR complex | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-08-25 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Atypical sideways recognition of CD1a by autoreactive gamma delta T cell receptors.
Nat Commun, 13, 2022
|
|
7RPP
 
 | | Crystal structure of human CEACAM1 with GFCC' and ABED face | | Descriptor: | 1,2-ETHANEDIOL, Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of human CEACAM1 oligomerization.
Commun Biol, 5, 2022
|
|
7FJD
 
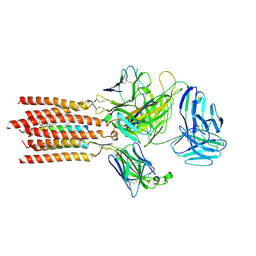 | | Cryo-EM structure of a membrane protein(WT) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJF
 
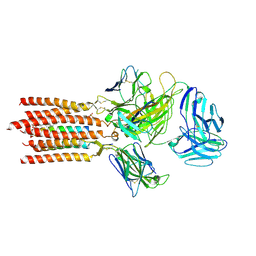 | | Cryo-EM structure of a membrane protein(CS) | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJE
 
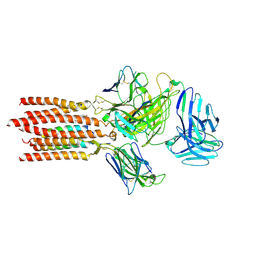 | | Cryo-EM structure of a membrane protein(LL) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7RNO
 
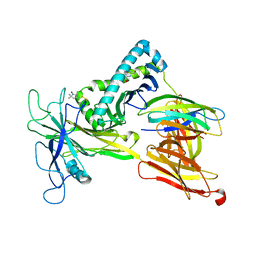 | | Model of the Ac-6-FP/hpMR1/bB2m/TAPBPR complex from integrated docking, NMR and restrained MD | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, N-(6-formyl-4-oxo-3,4-dihydropteridin-2-yl)acetamide, ... | | Authors: | McShan, A.C, Sgourakis, N.G. | | Deposit date: | 2021-07-29 | | Release date: | 2022-05-11 | | Last modified: | 2022-08-10 | | Method: | SOLUTION NMR | | Cite: | TAPBPR employs a ligand-independent docking mechanism to chaperone MR1 molecules.
Nat.Chem.Biol., 18, 2022
|
|
7RBU
 
 | | SARS-CoV-2 Spike in complex with PVI.V6-14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PVI.V6-14 Fab heavy chain, ... | | Authors: | Altomare, C.G, Bajic, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a Vaccine-Induced, Germline-Encoded Human Antibody Defines a Neutralizing Epitope on the SARS-CoV-2 Spike N-Terminal Domain.
Mbio, 13, 2022
|
|
7OUN
 
 | |
7N5P
 
 | | 6218 TCR in complex with H2-Db PA224-233 with a cysteine mutant | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 and T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5C
 
 | | 6218 TCR in complex with H2Db PA with an engineered TCR-pMHC disulfide bond | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 with T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-05 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N4K
 
 | | 6218 TCR in complex with H2-Db PA 224 | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 and T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-04 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7MU8
 
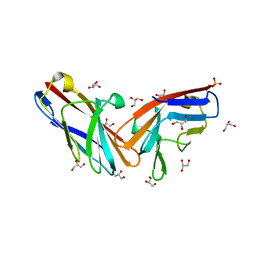 | | Structure of the minimally glycosylated human CEACAM1 N-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 1, GLYCEROL, ... | | Authors: | Belcher Dufrisne, M, Swope, N, Kieber, M, Yang, J.Y, Han, J, Li, J, Moremen, K.W, Prestegard, J.H, Columbus, L. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Human CEACAM1 N-domain dimerization is independent from glycan modifications.
Structure, 30, 2022
|
|
7EO0
 
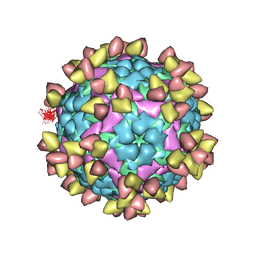 | | FOOT AND MOUTH DISEASE VIRUS O/TIBET/99-BOUND THE SINGLE CHAIN FRAGMEN ANTIBODY C4 | | Descriptor: | Ig heavy chain variable region, Ig lamda chain variable region, O/TIBET/99 VP1, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2021-04-21 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Two Cross-Protective Antigen Sites on Foot-and-Mouth Disease Virus Serotype O Structurally Revealed by Broadly Neutralizing Antibodies from Cattle.
J.Virol., 95, 2021
|
|
7ELX
 
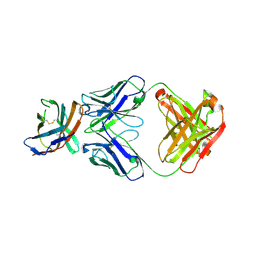 | | The crystal structure of CTLA-4 and Fab | | Descriptor: | Cytotoxic T-lymphocyte protein 4, heavy chain of Fab, light chain of Fab | | Authors: | Yu, X.J, Wang, L, Yu, C.F. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The crystal structure of CTLA-4 and Fab
To Be Published
|
|
