6N7Q
 
 | |
6MKT
 
 | | Photoactive Yellow Protein with 3-chlorotyrosine substituted at position 42 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
4DQO
 
 | | Crystal Structure of PG16 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 | | Descriptor: | 1FD6-V1V2 scaffold ZM109 HIV-1 strain, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG16 Fab Heavy Chain, ... | | Authors: | Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16.
Nat.Struct.Mol.Biol., 20, 2013
|
|
7BAD
 
 | | Crystal structure of PAFB in complex with p-sulfonato-calix[8]arene and zinc | | Descriptor: | Antifungal protein, PHOSPHATE ION, ZINC ION, ... | | Authors: | Guagnini, F, Huber, A, Alex, J.M, Marx, F, Crowley, P.B. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Porous assembly of an antifungal protein mediated by zinc and sulfonato-calix[8]arene.
J.Struct.Biol., 213, 2021
|
|
6MZ2
 
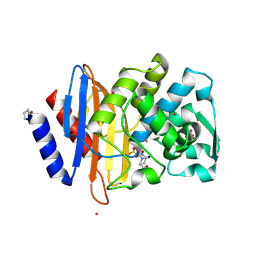 | |
7BAF
 
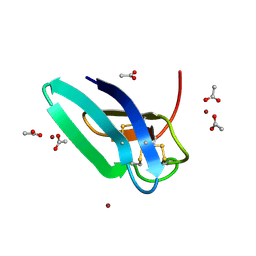 | | Crystal structure of PAFB in complex with zinc | | Descriptor: | ACETATE ION, Antifungal protein, ZINC ION | | Authors: | Guagnini, F, Huber, A, Alex, J.M, Marx, F, Crowley, P.B. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.123 Å) | | Cite: | Porous assembly of an antifungal protein mediated by zinc and sulfonato-calix[8]arene.
J.Struct.Biol., 213, 2021
|
|
7BAE
 
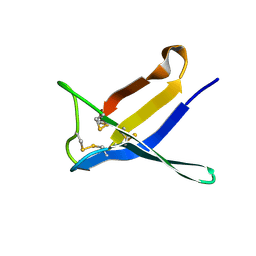 | | Crystal structure of PAFB | | Descriptor: | Antifungal protein | | Authors: | Guagnini, F, Huber, A, Alex, J.M, Marx, F, Crowley, P.B. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Porous assembly of an antifungal protein mediated by zinc and sulfonato-calix[8]arene.
J.Struct.Biol., 213, 2021
|
|
5J4P
 
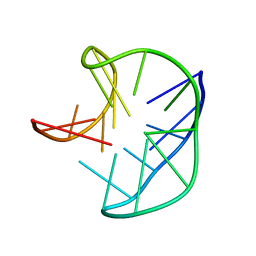 | |
5J4W
 
 | |
6MH9
 
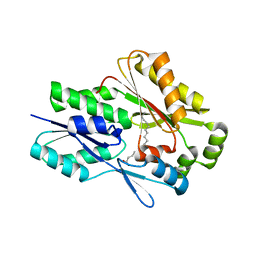 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein A121I mutant to 2.02 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B1 protein, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
6N9H
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Park, J, Baker, D. | | Deposit date: | 2018-12-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
5JQH
 
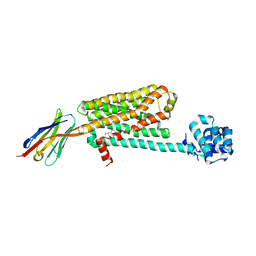 | | Structure of beta2 adrenoceptor bound to carazolol and inactive-state stabilizing nanobody, Nb60 | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, CHOLESTEROL, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Staus, D.P, Strachan, R.T, Manglik, A, Pani, B, Kahsai, A.W, Kim, T.H, Wingler, L.M, Ahn, S, Chatterjee, A, Masoudi, A, Kruse, A.C, Pardon, E, Steyaert, J, Weis, W.I, Prosser, R.S, Kobilka, B.K, Costa, T, Lefkowitz, R.J. | | Deposit date: | 2016-05-05 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation.
Nature, 535, 2016
|
|
5JYH
 
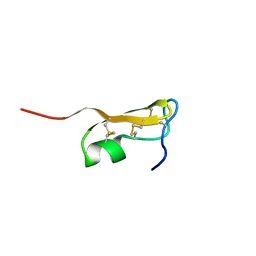 | |
5JP4
 
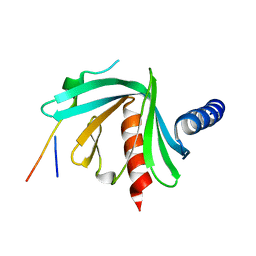 | |
6MIL
 
 | | Crystal structure of AF9 YEATS domain in complex with histone H3K9bu | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histone H3K9bu, MALONATE ION, ... | | Authors: | Vann, K.R, Klein, B.J, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6MMD
 
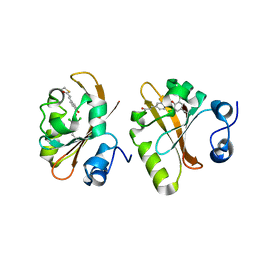 | | Photoactive Yellow Protein with 3,5-dichlorotyrosine substituted at position 42 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-30 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.228 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
6MXF
 
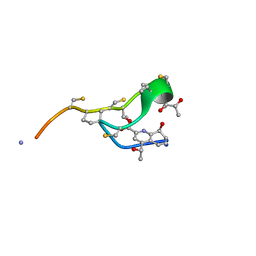 | | MicroED structure of thiostrepton at 1.9 A resolution | | Descriptor: | Thiostrepton | | Authors: | Jones, C.G, Martynowycz, M.W, Hattne, J, Fulton, T, Stoltz, B.M, Rodriguez, J.A, Nelson, H.M, Gonen, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.91 Å) | | Cite: | The CryoEM Method MicroED as a Powerful Tool for Small Molecule Structure Determination.
ACS Cent Sci, 4, 2018
|
|
6MZ1
 
 | |
5KF4
 
 | |
4DDM
 
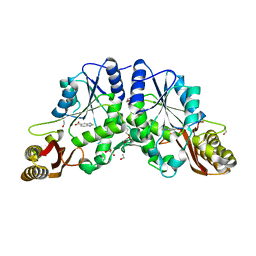 | | Pantothenate synthetase in complex with 2,1,3-benzothiadiazole-5-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 2,1,3-benzothiadiazole-5-carboxylic acid, ETHANOL, ... | | Authors: | Silvestre, H.L. | | Deposit date: | 2012-01-18 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6MNY
 
 | |
4CRG
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 6-carbamimidoyl-N-phenyl-4-(pyrimidin-2-ylamino)naphthalene-2-carboxamide, COAGULATION FACTOR XI, GLYCEROL, ... | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4CRE
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 6-chloro-4-methyl-1H-quinolin-2-one, COAGULATION FACTOR XI, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4CR5
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 6-chloroquinolin-2(1H)-one, COAGULATION FACTOR XIA, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-25 | | Release date: | 2015-02-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4CR9
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | 4-methylquinoline-2,6-diamine, COAGULATION FACTOR XI, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
