3B1O
 
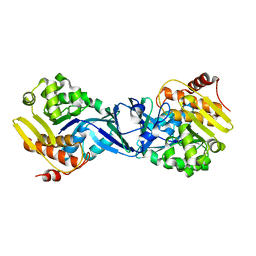 | | Structure of Burkholderia thailandensis nucleoside kinase (BthNK) in ligand-free form | | Descriptor: | Ribokinase, putative | | Authors: | Yasutake, Y, Ota, H, Hino, E, Sakasegawa, S, Tamura, T. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Burkholderia thailandensis nucleoside kinase: implications for the catalytic mechanism and nucleoside selectivity
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5NRY
 
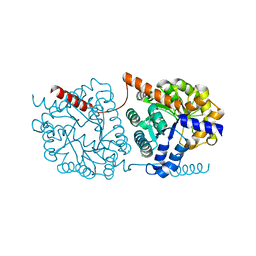 | | Cys-Gly dipeptidase GliJ in complex with Fe3+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, FE (III) ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
4RZF
 
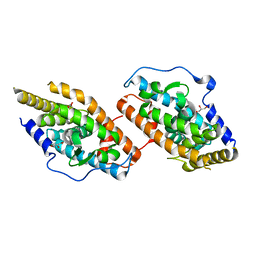 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, S441W mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
6WJN
 
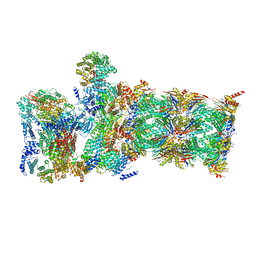 | |
3WKW
 
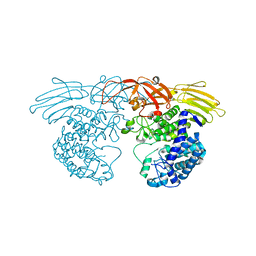 | | Crystal structure of GH127 beta-L-arabinofuranosidase HypBA1 from Bifidobacterium longum ligand free form | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Ito, T, Saikawa, K, Arakawa, T, Wakagi, T, Fujita, K. | | Deposit date: | 2013-11-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glycoside hydrolase family 127 beta-l-arabinofuranosidase from Bifidobacterium longum.
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4BUB
 
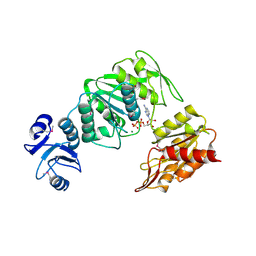 | | CRYSTAL STRUCTURE OF MURE LIGASE FROM THERMOTOGA MARITIMA IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--LD-LYSINE LIGASE | | Authors: | Favini-Stabile, S, Contreras-Martel, C, Thielens, N, Dessen, A. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mreb and Murg as Scaffolds for the Cytoplasmic Steps of Peptidoglycan Biosynthesis
Environ.Microbiol., 15, 2013
|
|
1ZUC
 
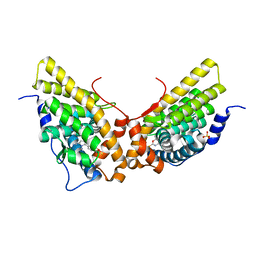 | | Progesterone receptor ligand binding domain in complex with the nonsteroidal agonist tanaproget | | Descriptor: | 5-(4,4-DIMETHYL-2-THIOXO-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-6-YL)-1-METHYL-1H-PYRROLE-2-CARBONITRILE, Progesterone receptor, SULFATE ION | | Authors: | Zhang, Z, Olland, A.M, Zhu, Y, Cohen, J, Berrodin, T, Chippari, S, Appavu, C, Li, S, Wilhem, J, Chopra, R, Fensome, A, Zhang, P, Wrobel, J, Unwalla, R.J, Lyttle, C.R, Winneker, R.C. | | Deposit date: | 2005-05-30 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and pharmacological properties of a potent and selective novel nonsteroidal progesterone receptor agonist tanaproget
J.Biol.Chem., 280, 2005
|
|
5FG1
 
 | |
4BUC
 
 | | CRYSTAL STRUCTURE OF MURD LIGASE FROM THERMOTOGA MARITIMA IN APO FORM | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Favini-Stabile, S, Contreras-Martel, C, Thielens, N, Dessen, A. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Mreb and Murg as Scaffolds for the Cytoplasmic Steps of Peptidoglycan Biosynthesis
Environ.Microbiol., 15, 2013
|
|
3QD6
 
 | | Crystal structure of the CD40 and CD154 (CD40L) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD40 ligand, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Lee, J.-O, Kim, Y.J, Song, D.H, Kim, H.M, Park, B.S. | | Deposit date: | 2011-01-18 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation
J.Biol.Chem., 286, 2011
|
|
3EJO
 
 | |
1B4M
 
 | | NMR STRUCTURE OF APO CELLULAR RETINOL-BINDING PROTEIN II, 24 STRUCTURES | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II | | Authors: | Lu, J, Lin, C.-L, Tang, C, Ponder, J.W, Kao, J.L.F, Cistola, D.P, Li, E. | | Deposit date: | 1998-12-23 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure and dynamics of rat apo-cellular retinol-binding protein II in solution: comparison with the X-ray structure.
J.Mol.Biol., 286, 1999
|
|
5VEY
 
 | | Solution NMR structure of histone H2A-H2B mono-ubiquitylated at H2A Lys15 in complex with RNF169 (653-708) | | Descriptor: | E3 ubiquitin-protein ligase RNF169, Histone H2B type 1-J,Histone H2A type 1-B/E, Polyubiquitin-B | | Authors: | Hu, Q, Botuyan, M.V, Cui, G, Mer, G. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Mechanisms of Ubiquitin-Nucleosome Recognition and Regulation of 53BP1 Chromatin Recruitment by RNF168/169 and RAD18.
Mol. Cell, 66, 2017
|
|
4RZE
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, L437W,D594E mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Fengwei, L, Xuyang, T, Anzhong, L, Li, L, Yuan, L, Hangzi, C, Qiao, W, Tianwei, L. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
1SXX
 
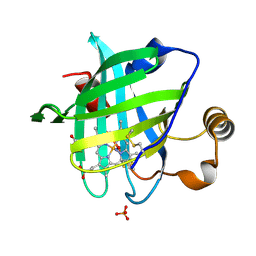 | | 1.0 A Crystal Structure of D129A/L130A Mutant of Nitrophorin 4 Complexed with Nitric Oxide | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
3WWR
 
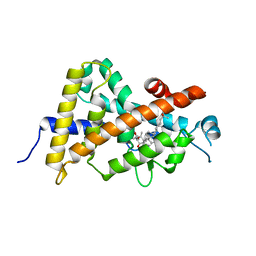 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 1-((((1S,2R,6R,Z)-2,6-dihydroxy-4-((E)-2-((1R,3aS,7aR)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methylhexahydro-1H-inden-4(2H)-ylidene)ethylidene)-3-methylenecyclohexyl)oxy)methyl)cyclopropanecarbonitrile | | Descriptor: | 1-({[(1R,2S,3R,5Z,7E,14beta,17alpha)-1,3,25-trihydroxy-9,10-secocholesta-5,7,10-trien-2-yl]oxy}methyl)cyclopropanecarbonitrile, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Synthesis and biological activities of vitamin D3 derivatives with cyanoalkyl side chain at C-2 position.
J.Steroid Biochem.Mol.Biol., 148, 2015
|
|
1MYG
 
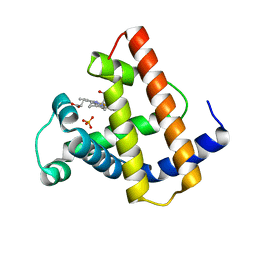 | | HIGH RESOLUTION X-RAY STRUCTURES OF PIG METMYOGLOBIN AND TWO CD3 MUTANTS MB(LYS45-> ARG) AND MB(LYS45-> SER) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Smerdon, S.J, Oldfield, T.J, Wilkinson, A.J, Dauter, Z, Petratos, K, Wilson, K.S. | | Deposit date: | 1992-02-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution X-ray structures of pig metmyoglobin and two CD3 mutants: Mb(Lys45----Arg) and Mb(Lys45----Ser).
Biochemistry, 31, 1992
|
|
4RZG
 
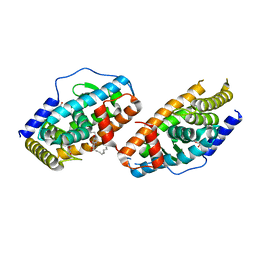 | | Crystal Structure Analysis of the DNPA-bounded NUR77 Ligand binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, pentyl (3,5-dihydroxy-2-nonanoylphenyl)acetate | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
3LN6
 
 | |
3THK
 
 | | Structure of SH3 chimera with a type II ligand linked to the chain C-terminal | | Descriptor: | BETA-MERCAPTOETHANOL, Proline-rich peptide, SULFATE ION, ... | | Authors: | Gabdulkhakov, A.G, Gushchina, L.V, Nikulin, A.D, Nikonov, S.V, Filimonov, V.V. | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structure of Spectrin SH3 Domain Fused with a Proline-Rich Peptide.
J.Biomol.Struct.Dyn., 29, 2011
|
|
1HNF
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR REGION OF THE HUMAN CELL ADHESION MOLECULE CD2 AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD2, SODIUM ION | | Authors: | Bodian, D.L, Jones, E.Y, Harlos, K, Stuart, D.I, Davis, S.J. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 A resolution.
Structure, 2, 1994
|
|
4WI1
 
 | |
4ZLG
 
 | | Cellobionic acid phosphorylase - gluconic acid complex | | Descriptor: | CHLORIDE ION, D-gluconic acid, D-glucono-1,5-lactone, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
3EJT
 
 | | Golgi alpha-Mannosidase II in complex with 5-substituted swainsonine analog:(5R)-5-[2'-(4-tert-butylphenyl)ethyl]-swainsonine | | Descriptor: | (1S,2R,5R,8R,8aR)-5-[2-(4-tert-butylphenyl)ethyl]octahydroindolizine-1,2,8-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Shea, K, Rose, D.R. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Investigation of the Binding of 5-Substituted Swainsonine Analogues to Golgi alpha-Mannosidase II.
Chembiochem, 11, 2010
|
|
3EJR
 
 | | Golgi alpha-Mannosidase II in complex with 5-substitued swainsonine analog: (5R)-5-[2'-oxo-2'-(4-tert-butylphenyl)ethyl]-swainsonine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-(4-tert-butylphenyl)-2-[(1S,2R,5R,8R,8aR)-1,2,8-trihydroxyoctahydroindolizin-5-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Shea, K, Rose, D.R. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural Investigation of the Binding of 5-Substituted Swainsonine Analogues to Golgi alpha-Mannosidase II.
Chembiochem, 11, 2010
|
|
