3SD2
 
 | |
3SIG
 
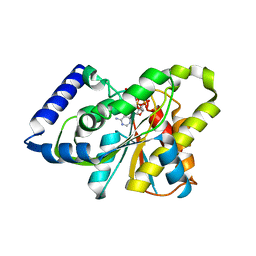 | | The X-ray crystal structure of poly(ADP-ribose) glycohydrolase (PARG) bound to ADP-ribose from Thermomonospora curvata | | Descriptor: | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE, poly(ADP-ribose) glycohydrolase | | Authors: | Leys, D, Dunstan, M.S. | | Deposit date: | 2011-06-18 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase.
Nature, 477, 2011
|
|
3SDI
 
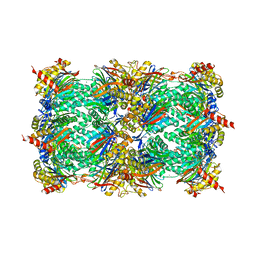 | | Structure of yeast 20S open-gate proteasome with Compound 20 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N~4~-(2,2-dimethylpropyl)-N~1~-{(2S)-1-[(4-methylbenzyl)amino]-1-oxo-4-phenylbutan-2-yl}-N~2~-[(5-methyl-1,2-oxazol-3-yl)carbonyl]-L-aspartamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-06-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3SIW
 
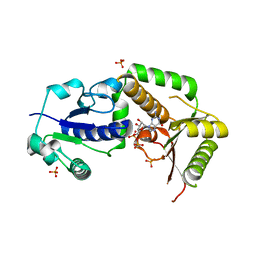 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase co-crystallized with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SFG
 
 | |
3SJH
 
 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-29) of drosophila Ciboulot and the C-terminal domain (residues 18-44) of bovine Thymosin-beta4, bound to G-actin-ATP-Latrunculin A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-06-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
3SJR
 
 | |
3SK4
 
 | |
3SHP
 
 | | Crystal structure of putative acetyltransferase from Sphaerobacter thermophilus DSM 20745 | | Descriptor: | Putative acetyltransferase Sthe_0691, S,R MESO-TARTARIC ACID | | Authors: | Chang, C, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of putative acetyltransferase from Sphaerobacter thermophilus DSM 20745
To be Published
|
|
3SI2
 
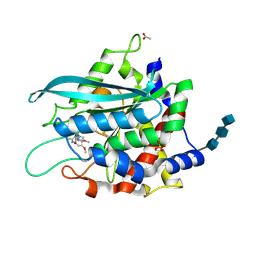 | | Structure of glycosylated murine glutaminyl cyclase in presence of the inhibitor PQ50 (PDBD150) | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Parthier, C, Carrillo, D, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
3SIE
 
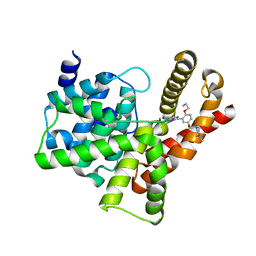 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SMB
 
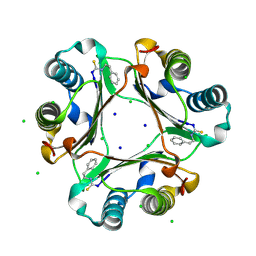 | |
3SME
 
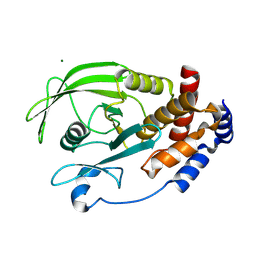 | | Structure of PTP1B inactivated by H2O2/bicarbonate | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Tanner, J.J, Singh, H. | | Deposit date: | 2011-06-27 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Biological Buffer Bicarbonate/CO(2) Potentiates H(2)O(2)-Mediated Inactivation of Protein Tyrosine Phosphatases.
J.Am.Chem.Soc., 133, 2011
|
|
3SK9
 
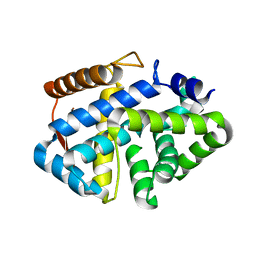 | | Crystal structure of the Thermus thermophilus cas3 HD domain | | Descriptor: | Putative uncharacterized protein TTHB187 | | Authors: | Bailey, S, Mulepati, S. | | Deposit date: | 2011-06-22 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Analysis of Nuclease Domain of Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-associated Protein 3 (Cas3).
J.Biol.Chem., 286, 2011
|
|
3SNO
 
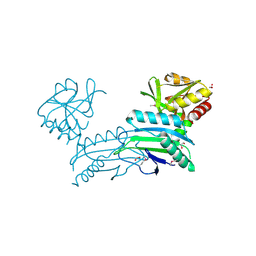 | |
3SO6
 
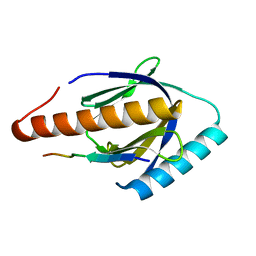 | |
3S4P
 
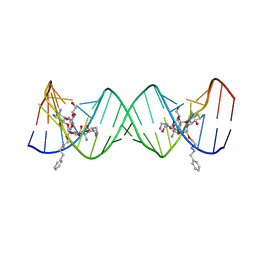 | | Crystal structure of the bacterial ribosomal decoding site complexed with an amphiphilic paromomycin O2''-ether analogue | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-2-O-{2-[(2-phenylethyl)amino]ethyl}-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, RNA (5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Szychowski, J, Kondo, J, Zahr, O, Auclair, K, Westhof, E, Hanessian, S, Keillor, J.W. | | Deposit date: | 2011-05-20 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Inhibition of aminoglycoside-deactivating enzymes APH(3')-IIIa and AAC(6')-Ii by amphiphilic paromomycin O2''-ether analogues
Chemmedchem, 6, 2011
|
|
3S5R
 
 | |
3S9T
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-{[(4,6-dimethyl-2-pyrimidinyl)sulfanyl]acetyl}benzenesulfonamide | | Descriptor: | 2-chloro-5-{[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, BICINE, Carbonic anhydrase 2, ... | | Authors: | Grazulis, S, Manakova, E, Tamulaitiene, G. | | Deposit date: | 2011-06-02 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of [(2-pyrimidinylthio)acetyl]benzenesulfonamides as inhibitors of human carbonic anhydrases.
Eur.J.Med.Chem., 51, 2012
|
|
3SA2
 
 | |
3SAA
 
 | | Crystal structure of Wild-type HIV-1 protease in complex With AF77 | | Descriptor: | N-[(2S,3R)-4-{(cyclohexylmethyl)[(4-methoxyphenyl)sulfonyl]amino}-3-hydroxy-1-phenylbutan-2-yl]-3-hydroxybenzamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SB6
 
 | | Cu-mediated Dimer of T4 Lysozyme D61H/K65H/R76H/R80H by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Lysozyme | | Authors: | Soriaga, A.B, Laganowsky, A, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3S6E
 
 | |
3SA3
 
 | | Crystal structure of wild-type HIV-1 protease in complex with AG23 | | Descriptor: | ACETATE ION, N~2~-acetyl-N-[(2S,3R)-4-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]-L-isoleucinamide, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SGW
 
 | |
